官网MetaboAnalyst
一、安装rtools并设置系统环境path
1.首先安装rtools
参考帖子:
Rtools下载与安装(win10) - 知乎 (zhihu.com)
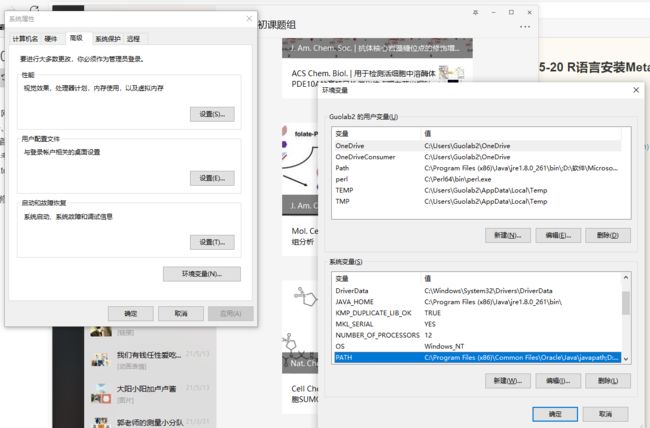
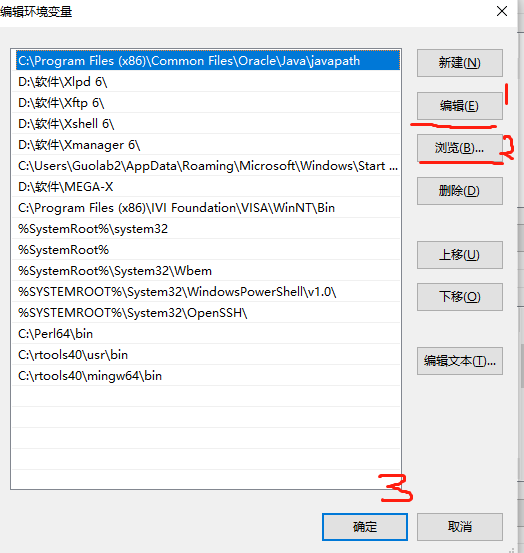
2.修改path
3.重新打开Rstudio
After installation is complete, you need to perform one more step to be able to compile R packages: you need to put the location of the Rtools make utilities (bash, make, etc) on the PATH. The easiest way to do so is create a text file .Renviron in your Documents folder which contains the following line:
PATH="${RTOOLS40_HOME}\usr\bin;${PATH}"
You can do this with a text editor, or from R like so (note that in R code you need to escape backslashes):
writeLines('PATH="${RTOOLS40_HOME}\\usr\\bin;${PATH}"', con = "~/.Renviron")
Now restart R, and verify that make can be found, which should show the path to your Rtools installation.
Sys.which("make")
## "C:\\rtools40\\usr\\bin\\make.exe"
If this works, you can try to install an R package from source:
install.packages("jsonlite", type = "source")
参考
https://cran.r-project.org/bin/windows/Rtools/
4.出现了Permission denied的问题
查看R语言-解决problem copying rlang.dll: Permission denied的问题 - (jianshu.com)
R包安装Permission denied 问题解决_pythonic生物人的博客-CSDN博客这两个帖子进行修改,并把r-4.0下边的切换到路径D:\learning\R\R-4.0.2\library
将00LOCK\digest\及digest文件夹全部删除,重新安装即可。
二、安装
writeLines('PATH="${RTOOLS40_HOME}\\usr\\bin;${PATH}"', con = "~/.Renviron")
Sys.which("make")
install.packages("jsonlite", type = "source")
library(jsonlite)
metanr_packages <- function(){
metr_pkgs <- c("impute", "pcaMethods", "globaltest", "GlobalAncova", "Rgraphviz", "preprocessCore", "genefilter", "SSPA", "sva", "limma", "KEGGgraph", "siggenes","BiocParallel", "MSnbase", "multtest","RBGL","edgeR","fgsea","devtools","crmn")
list_installed <- installed.packages()
new_pkgs <- subset(metr_pkgs, !(metr_pkgs %in% list_installed[, "Package"]))
if(length(new_pkgs)!=0){if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install(new_pkgs)
print(c(new_pkgs, " packages added..."))
}
if((length(new_pkgs)<1)){
print("No new packages added...")
}
}
metanr_packages()
library(pacman)
pacman::p_load(c("impute", "pcaMethods", "globaltest", "GlobalAncova", "Rgraphviz", "preprocessCore", "genefilter", "SSPA", "sva", "limma", "KEGGgraph", "siggenes","BiocParallel", "MSnbase", "multtest","RBGL","edgeR","fgsea"))
##利用pacman后面安装一直出问题,后面进行单独的library后,有些包需要更新就更新,有些错误就看看如何解决错误,直到所有的library都没有问题后进行下载
library(pcaMethods)
library(globaltest)
library(GlobalAncova)
library(Rgraphviz)
library(preprocessCore)
library(genefilter)
# if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
#
# BiocManager::install("SSPA",force = TRUE)
library(SSPA)
library(sva)
library(limma)
library(KEGGgraph)
library(siggenes)
library(BiocParallel)
library(MSnbase)
library(multtest)
library(RBGL)
library(edgeR)
library(fgsea)
BiocManager::install("glasso",force = TRUE)
BiocManager::install("rmarkdown",force = TRUE)
BiocManager::install("Matrix",force = TRUE)
library(glasso)
library(remarkdown)
library(devtools)
# Step 2: Install MetaboAnalystR without documentation
devtools::install_github("xia-lab/MetaboAnalystR", build = TRUE, build_vignettes = FALSE)
library("MetaboAnalystR")