MachineLearning 10. 癌症诊断机器学习之神经网络(Neural Network)
点击关注,桓峰基因
桓峰基因
生物信息分析,SCI文章撰写及生物信息基础知识学习:R语言学习,perl基础编程,linux系统命令,Python遇见更好的你
99篇原创内容
公众号
通过乳腺癌是数据我们利用不同的机器学习算法,不断的解开机器学习的神秘面纱,使得这种AI技术能够让医学更加适用,不再是一件神秘的算法,而已都能接受的方法而已!这期就来说说神经网络建模对乳腺癌的诊断效果!!
前 言
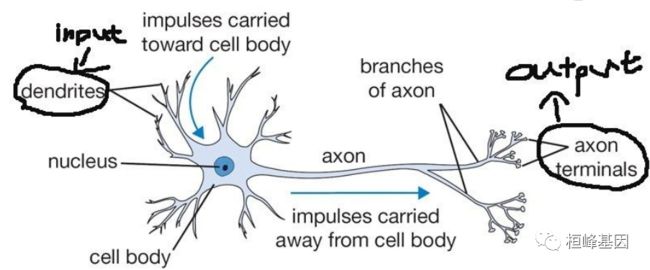
高中生物基本忘记得差不多了,不过依稀还记得我们大脑对外界的大概反应过程可以描述为外界的刺激信号传进某部分的神经元系统,信号经过神经元一层层地传递下去,最终在某部分的神经元系统产生脉冲信号,驱使身体的某个部位作出反应。神经元的结构如下:输入信号从突触(dendrites)进入后,转化为化学信号,经过神经元细胞一系列的处理,最终在轴突末梢(axon terminals)转换为输出信号。
基本原理
在下图这个简单的网络中,输入(又称协变量)由两个节点(又称神经元)组成。标 有1的神经元代表一个常数,更确切地说,是截距。XI衣公化黑tI’WoN重,会和输入节点的值相乘。输入节点的值在与W相乘后传递到隐臧力扁。P以月多个隐藏节点,但是工作原理和一个隐藏节点没有什么不同。在隐藏节点H1中,所有 加权后的输入值被加总。因为截距为1,所以这个输入节点的值就是权重W1。加总后 的值通过激活函数进行转换,将输入信号转换为输出信号。在这个简里例子中,HL是唯一的隐藏节点,它的值被乘以W3,成为响应变量Y的估计值。这就是算法的前馈 部分。要完成这个循环,或者用神经网络中的说法,完成一次完整的“训练”,还要进行反向传播过程,基于学习到的知识来训练模型。为了初始化区同传播过程,需I丁坝失函数确定误差,损失函数可以是误差平方总和,也可以是交叉嫡,或者其他形式。因为权重W1和W2最初被设定为[-1,1]之间的随机数,所以初始的误差可能会很大。风向传播时,要改变权重值以使损失函数中的误差最小。下页的图描述了算法的反向传 播部分。这样就完成了一次完整的训练。这个过程不断继续,使用梯度下降方法可减小误差,直到算法收敛于误差最小值或者达到预先设定的训练次数。如果假设本例中的激活函数是简单线性的,那么结果为Y=W3(W1(1)+ W2(X1))。如果你增加了大量的输入神经元,或者在隐藏节点中增加多个神经元,甚至增加多个隐藏节点,神经网络会变得非常复杂。请一定注意,一个神经元的输出会连接到所有后继的神经元,并将权重分配给所有连接,这样会大大增加模型的复杂性。增加隐藏节点和提高隐藏节点中神经元的数量不会像我们希望的那样改善模型的性能。于是,深度学习得以发展,它可以部分放松所有神经元都要连接的要求。有很多种激活函数可供使用,其中包括一个简单线性函数,还有用于分类问题的sigmoid函数,它是Logistic函数的一种特殊形式。当输出变量 变量(0或1)时,可以使用阈值函数。其他常用的激活函数还有Rectifier、Maxout以及双曲正切函数(tanh)。
实例解析
这期我们仍然使用乳腺癌的数据集,对比之前的机器学习算法,探究神经网络在临床上的应用。
1. 软件安装
我们这里使用软件包neuralnet,配套有绘图等函数,另外还有caret用于机器学习非常方便的软件包,软件安装并加载,如下:
if (!require(neuralnet)) install.packages("neuralnet")
if (!require(vcd)) install.packages("vcd")
if (!require(caret)) install.packages("caret")
library(caret)
library(neuralnet)
library(vcd)
2. 数据读取
数据来源《机器学习与R语言》书中,具体来自UCI机器学习仓库。地址:http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/ 下载wbdc.data和wbdc.names这两个数据集,数据经过整理,成为面板数据。查看数据结构,其中第一列为id列,无特征意义,需要删除。第二列diagnosis为响应变量(B,M),字符型,一般在R语言中分类任务都要求响应变量为因子类型,因此需要做数据类型转换。剩余的为预测变量,数值类型。查看数据维度,568个样本,32个特征(包括响应特征)。
BreastCancer <- read.csv("wisc_bc_data.csv", stringsAsFactors = FALSE)
dim(BreastCancer)
## [1] 568 32
table(BreastCancer$diagnosis)
##
## B M
## 357 211
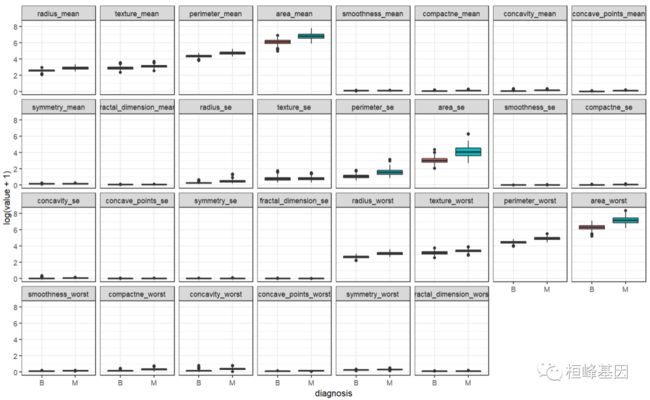
3. 数据分布
比较恶性和良性之间的差距
bc <- BreastCancer[, -1]
library(reshape2)
bc.melt <- melt(bc, id.var = "diagnosis")
head(bc.melt)
## diagnosis variable value
## 1 M radius_mean 20.57
## 2 M radius_mean 19.69
## 3 M radius_mean 11.42
## 4 M radius_mean 20.29
## 5 M radius_mean 12.45
## 6 M radius_mean 18.25
ggplot(data = bc.melt, aes(x = diagnosis, y = log(value + 1), fill = diagnosis)) +
geom_boxplot() + theme_bw() + facet_wrap(~variable, ncol = 8)
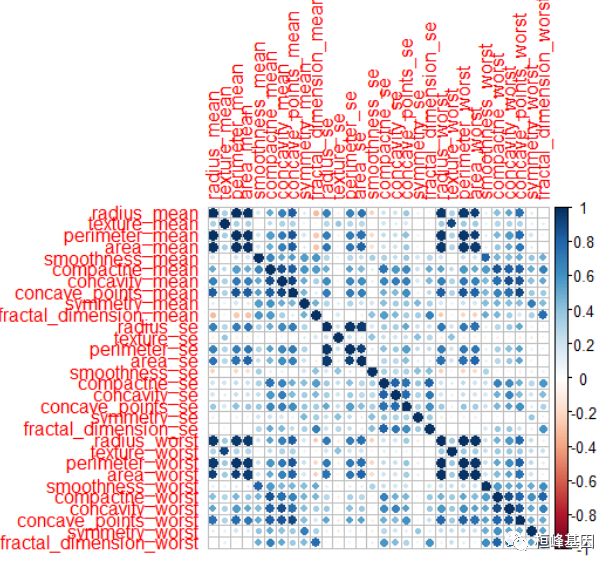
比较变量之间的相关性,如下:
corrplot::corrplot(cor(bc[, -1]))
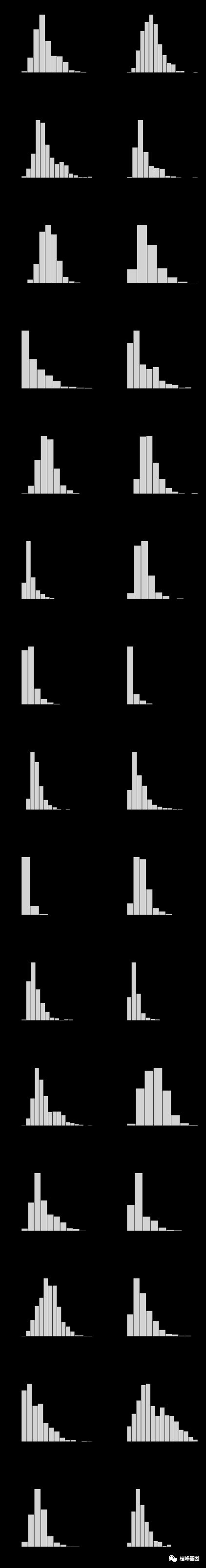
乳腺癌数据集分布的直方图,如下:
library(tidyverse)
data <- select(BreastCancer, -1) %>%
mutate_at("diagnosis", as.factor)
n = names(data[, 2:31])
pdf("hist.pdf", width = 6, height = 45)
par(mfrow = c(15, 2))
for (i in1:30) {
x = unlist(data.frame(data[n[i]]))
hist(x, main = paste("Histogram of ", n[i], sep = ""), xlab = n[i])
}
dev.off()
## png
## 2
新增分组列
根据数据集在diagnosis列取值不同,为数据集新增CancerB,CancerM数据列,如下:
Cancer <- data$diagnosis
Cancer <- as.data.frame(Cancer)
Cancer <- with(Cancer, data.frame(model.matrix(~Cancer + 0)))
data <- as.data.frame(cbind(data, Cancer))
head(data, 2)
## diagnosis radius_mean texture_mean perimeter_mean area_mean smoothness_mean
## 1 M 20.57 17.77 132.9 1326 0.08474
## 2 M 19.69 21.25 130.0 1203 0.10960
## compactne_mean concavity_mean concave_points_mean symmetry_mean
## 1 0.07864 0.0869 0.07017 0.1812
## 2 0.15990 0.1974 0.12790 0.2069
## fractal_dimension_mean radius_se texture_se perimeter_se area_se
## 1 0.05667 0.5435 0.7339 3.398 74.08
## 2 0.05999 0.7456 0.7869 4.585 94.03
## smoothness_se compactne_se concavity_se concave_points_se symmetry_se
## 1 0.005225 0.01308 0.01860 0.01340 0.01389
## 2 0.006150 0.04006 0.03832 0.02058 0.02250
## fractal_dimension_se radius_worst texture_worst perimeter_worst area_worst
## 1 0.003532 24.99 23.41 158.8 1956
## 2 0.004571 23.57 25.53 152.5 1709
## smoothness_worst compactne_worst concavity_worst concave_points_worst
## 1 0.1238 0.1866 0.2416 0.186
## 2 0.1444 0.4245 0.4504 0.243
## symmetry_worst fractal_dimension_worst CancerB CancerM
## 1 0.2750 0.08902 0 1
## 2 0.3613 0.08758 0 1
4. 数据分割
当我们只有一套数据的时候,可以将数据分为训练集和测试集,具体怎么分割可以看公众号的专题:Topic 5. 样本量确定及分割我们将整个数据进行分割,分为训练集和测试集,并保证其正负样本的比例,如下:
# install.packages('sampling')
library(sampling)
set.seed(123)
# 每层抽取70%的数据
train_id <- strata(data, "diagnosis", size = rev(round(table(data$diagnosis) * 0.7)))$ID_unit
# 训练数据
train_data <- data[train_id, -1]
# 测试数据
test_data <- data[-train_id, -1]
# 查看训练、测试数据中正负样本比例
prop.table(table(train_data$CancerB))
##
## 0 1
## 0.3718593 0.6281407
prop.table(table(test_data$CancerB))
##
## 0 1
## 0.3705882 0.6294118
5. 实例操作
我们利用分割好的乳腺癌数据集进行神经网络模型的构建,并对其结果进行验证,绘制神经网络图。
构建神经网络
调用neuralnet函数创建一个包含3个隐藏层的神经网络,训练结果有可能随机发生变化,所以得到的结果可能不同,可以开始指定seed值使得每次训练返回相同的值。
neuralnet函数参数说明:
formula :公式;
data :建模的数据;
hidden :每个隐藏层的单元个数;
threshold :误差函数的停止阈值;
stepmax :最大迭代次数;
rep :神经网络训练的重复次数;
startweights :初始权值,不会随机初始权值了;
learningrate.limit :学习率的上下限,只针对学习函数为RPROP和GRPROP;
learningrate.factor :同上,不过可以是针对多个;
learningrate :算法的学习速率,只针对BP算法;
lifesign :神经网络计算过程中打印多少函数{none、minimal、full};
algorithm :计算神经网络的算法{ backprop , rprop+ , rprop- , sag , slr };
err.fct :计算误差,’{sse,se};
act.fct :激活函数,{logistic,tanh};
linear.output :是否线性输出,即是回归还是分类;
exclude :一个用来指定在计算中将要排除的权重的向量或矩阵,如果给的是一个向量,则权重的位置很明确,如果是一个n*3的矩阵,则会排除n个权重,第一列表示层数,第二列,第三列分别表示权重的输入单元和输出单元;
constant.weights :指定训练过程中不需要训练的权重,在训练中看做固定值;
likelihood :逻辑值,如果损失函数是负对数似然函数,那么信息标准AIC和BIC将会被计算。
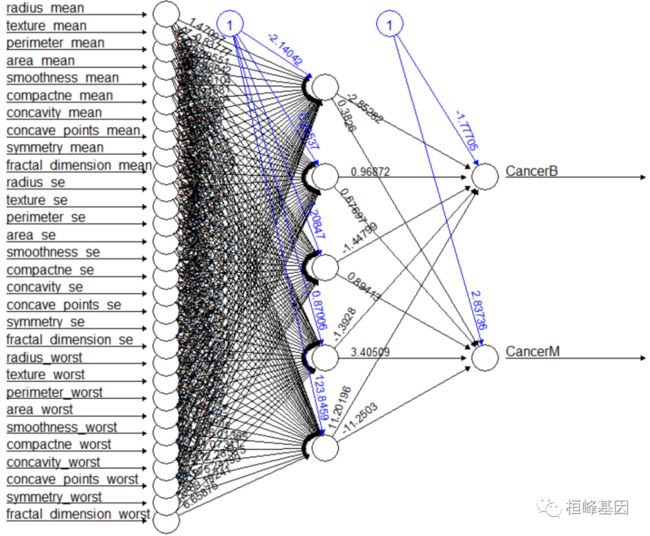
这里构建神经网络我们使用的是neuralnet函数,做左边是变量名,每条线上的数值是权重值,隐藏层为3,如下:
set.seed(123)
Col = names(train_data[, 1:(ncol(train_data) - 2)])
f = as.formula(paste("CancerB + CancerM ~", paste(Col, collapse = " + ")))
net = neuralnet(f, data = train_data, hidden = 5, linear.output = FALSE)
plot(net)
输出构建好的神经网络模型的结果矩阵result.matrix,我们发现残差error=1.347026e+01,非常的小,说明结果不错。
net$result.matrix
## [,1]
## error 1.347026e+01
## reached.threshold 9.215291e-03
## steps 1.788000e+03
## Intercept.to.1layhid1 6.643849e+01
## radius_mean.to.1layhid1 6.627828e-02
## texture_mean.to.1layhid1 1.861271e+00
## perimeter_mean.to.1layhid1 3.667256e-01
## area_mean.to.1layhid1 3.571613e-01
## smoothness_mean.to.1layhid1 3.079611e+01
## compactne_mean.to.1layhid1 -2.002705e+01
## concavity_mean.to.1layhid1 -1.142198e+02
## concave_points_mean.to.1layhid1 -1.348972e+02
## symmetry_mean.to.1layhid1 3.074433e+00
## fractal_dimension_mean.to.1layhid1 4.820844e+01
## radius_se.to.1layhid1 3.712372e+01
## texture_se.to.1layhid1 -2.441092e+00
## perimeter_se.to.1layhid1 6.886018e+00
## area_se.to.1layhid1 -2.337285e-01
## smoothness_se.to.1layhid1 -4.393309e+01
## compactne_se.to.1layhid1 2.297421e+01
## concavity_se.to.1layhid1 3.673323e+01
## concave_points_se.to.1layhid1 1.129134e+00
## symmetry_se.to.1layhid1 -2.355318e+01
## fractal_dimension_se.to.1layhid1 6.458040e+00
## radius_worst.to.1layhid1 7.848375e-02
## texture_worst.to.1layhid1 -8.395178e-01
## perimeter_worst.to.1layhid1 -4.326636e-01
## area_worst.to.1layhid1 -3.348451e-01
## smoothness_worst.to.1layhid1 1.194755e+01
## compactne_worst.to.1layhid1 -1.978799e+01
## concavity_worst.to.1layhid1 -4.216651e+01
## concave_points_worst.to.1layhid1 -2.935827e+01
## symmetry_worst.to.1layhid1 -2.536716e+01
## fractal_dimension_worst.to.1layhid1 6.234373e+00
## Intercept.to.1layhid2 -2.950715e-01
## radius_mean.to.1layhid2 8.951257e-01
## texture_mean.to.1layhid2 8.781335e-01
## perimeter_mean.to.1layhid2 8.215811e-01
## area_mean.to.1layhid2 6.886403e-01
## smoothness_mean.to.1layhid2 5.539177e-01
## compactne_mean.to.1layhid2 -6.191171e-02
## concavity_mean.to.1layhid2 -3.059627e-01
## concave_points_mean.to.1layhid2 -3.804710e-01
## symmetry_mean.to.1layhid2 -6.947070e-01
## fractal_dimension_mean.to.1layhid2 -2.079173e-01
## radius_se.to.1layhid2 -1.265396e+00
## texture_se.to.1layhid2 2.168956e+00
## perimeter_se.to.1layhid2 1.207962e+00
## area_se.to.1layhid2 -1.123109e+00
## smoothness_se.to.1layhid2 -4.028848e-01
## compactne_se.to.1layhid2 -4.666554e-01
## concavity_se.to.1layhid2 7.799651e-01
## concave_points_se.to.1layhid2 -8.336907e-02
## symmetry_se.to.1layhid2 2.533185e-01
## fractal_dimension_se.to.1layhid2 -2.854676e-02
## radius_worst.to.1layhid2 -4.287046e-02
## texture_worst.to.1layhid2 1.368602e+00
## perimeter_worst.to.1layhid2 -2.257710e-01
## area_worst.to.1layhid2 1.516471e+00
## smoothness_worst.to.1layhid2 -1.548753e+00
## compactne_worst.to.1layhid2 5.846137e-01
## concavity_worst.to.1layhid2 1.238542e-01
## concave_points_worst.to.1layhid2 2.159416e-01
## symmetry_worst.to.1layhid2 3.796395e-01
## fractal_dimension_worst.to.1layhid2 -5.023235e-01
## Intercept.to.1layhid3 -3.332074e-01
## radius_mean.to.1layhid3 -1.018575e+00
## texture_mean.to.1layhid3 -1.071791e+00
## perimeter_mean.to.1layhid3 3.035286e-01
## area_mean.to.1layhid3 4.482098e-01
## smoothness_mean.to.1layhid3 5.300423e-02
## compactne_mean.to.1layhid3 9.222675e-01
## concavity_mean.to.1layhid3 2.050085e+00
## concave_points_mean.to.1layhid3 -4.910312e-01
## symmetry_mean.to.1layhid3 -2.309169e+00
## fractal_dimension_mean.to.1layhid3 1.005739e+00
## radius_se.to.1layhid3 -7.092008e-01
## texture_se.to.1layhid3 -6.880086e-01
## perimeter_se.to.1layhid3 1.025571e+00
## area_se.to.1layhid3 -2.847730e-01
## smoothness_se.to.1layhid3 -1.220718e+00
## compactne_se.to.1layhid3 1.813035e-01
## concavity_se.to.1layhid3 -1.388914e-01
## concave_points_se.to.1layhid3 5.764186e-03
## symmetry_se.to.1layhid3 3.852804e-01
## fractal_dimension_se.to.1layhid3 -3.706600e-01
## radius_worst.to.1layhid3 6.443765e-01
## texture_worst.to.1layhid3 -2.204866e-01
## perimeter_worst.to.1layhid3 3.317820e-01
## area_worst.to.1layhid3 1.096839e+00
## smoothness_worst.to.1layhid3 4.351815e-01
## compactne_worst.to.1layhid3 -3.259316e-01
## concavity_worst.to.1layhid3 1.148808e+00
## concave_points_worst.to.1layhid3 9.935039e-01
## symmetry_worst.to.1layhid3 5.483970e-01
## fractal_dimension_worst.to.1layhid3 2.387317e-01
## Intercept.to.1layhid4 -6.279061e-01
## radius_mean.to.1layhid4 1.360652e+00
## texture_mean.to.1layhid4 -6.002596e-01
## perimeter_mean.to.1layhid4 2.187333e+00
## area_mean.to.1layhid4 1.532611e+00
## smoothness_mean.to.1layhid4 -2.357004e-01
## compactne_mean.to.1layhid4 -1.026421e+00
## concavity_mean.to.1layhid4 -7.104066e-01
## concave_points_mean.to.1layhid4 2.568837e-01
## symmetry_mean.to.1layhid4 -2.466919e-01
## fractal_dimension_mean.to.1layhid4 -3.475426e-01
## radius_se.to.1layhid4 -9.516186e-01
## texture_se.to.1layhid4 -4.502772e-02
## perimeter_se.to.1layhid4 -7.849045e-01
## area_se.to.1layhid4 -1.667942e+00
## smoothness_se.to.1layhid4 -3.802265e-01
## compactne_se.to.1layhid4 9.189966e-01
## concavity_se.to.1layhid4 -5.753470e-01
## concave_points_se.to.1layhid4 6.079643e-01
## symmetry_se.to.1layhid4 -1.617883e+00
## fractal_dimension_se.to.1layhid4 -5.556197e-02
## radius_worst.to.1layhid4 5.194072e-01
## texture_worst.to.1layhid4 3.011534e-01
## perimeter_worst.to.1layhid4 1.056762e-01
## area_worst.to.1layhid4 -6.407060e-01
## smoothness_worst.to.1layhid4 -8.497043e-01
## compactne_worst.to.1layhid4 -1.024129e+00
## concavity_worst.to.1layhid4 1.176466e-01
## concave_points_worst.to.1layhid4 -9.474746e-01
## symmetry_worst.to.1layhid4 -4.905574e-01
## fractal_dimension_worst.to.1layhid4 -2.560922e-01
## Intercept.to.1layhid5 1.843862e+00
## radius_mean.to.1layhid5 -6.519499e-01
## texture_mean.to.1layhid5 2.353866e-01
## perimeter_mean.to.1layhid5 7.796085e-02
## area_mean.to.1layhid5 -9.618566e-01
## smoothness_mean.to.1layhid5 -7.130809e-02
## compactne_mean.to.1layhid5 1.444551e+00
## concavity_mean.to.1layhid5 4.515041e-01
## concave_points_mean.to.1layhid5 4.123292e-02
## symmetry_mean.to.1layhid5 -4.224968e-01
## fractal_dimension_mean.to.1layhid5 -2.053247e+00
## radius_se.to.1layhid5 1.131337e+00
## texture_se.to.1layhid5 -1.460640e+00
## perimeter_se.to.1layhid5 7.399475e-01
## area_se.to.1layhid5 1.909104e+00
## smoothness_se.to.1layhid5 -1.443893e+00
## compactne_se.to.1layhid5 7.017843e-01
## concavity_se.to.1layhid5 -2.621975e-01
## concave_points_se.to.1layhid5 -1.572144e+00
## symmetry_se.to.1layhid5 -1.514668e+00
## fractal_dimension_se.to.1layhid5 -1.601536e+00
## radius_worst.to.1layhid5 -5.309065e-01
## texture_worst.to.1layhid5 -1.461756e+00
## perimeter_worst.to.1layhid5 6.879168e-01
## area_worst.to.1layhid5 2.100109e+00
## smoothness_worst.to.1layhid5 -1.287030e+00
## compactne_worst.to.1layhid5 7.877388e-01
## concavity_worst.to.1layhid5 7.690422e-01
## concave_points_worst.to.1layhid5 3.322026e-01
## symmetry_worst.to.1layhid5 -1.008377e+00
## fractal_dimension_worst.to.1layhid5 -1.194526e-01
## Intercept.to.CancerB -1.249642e+00
## 1layhid1.to.CancerB 6.959205e+00
## 1layhid2.to.CancerB -1.341685e+00
## 1layhid3.to.CancerB 7.726790e-03
## 1layhid4.to.CancerB -1.343827e+00
## 1layhid5.to.CancerB 8.346487e-02
## Intercept.to.CancerM -8.225165e-01
## 1layhid1.to.CancerM -6.957986e+00
## 1layhid2.to.CancerM 3.467700e+00
## 1layhid3.to.CancerM -1.901970e-01
## 1layhid4.to.CancerM 5.248881e-01
## 1layhid5.to.CancerM 8.632302e-01
调用head函数,返回network模型的犬种第一项。原理:神经网络中的神经元是相互连接的,不同神经元之间连接作用的强弱被称为节点的链接权重。
隐藏神经元的数量越多,error值越小,越好,计算越复杂···
从输出结果可知,整个训练执行了398*60步,终止条件为误差函数的绝对偏导数小于0.01(reached.threshold),误差依然值的计算采用AIC准则。
从输出结果可知,模型的权值范围在-58到58之间;第一层隐含网络的截距分别为-66.4,-0.3,-0.33,-0.63, 1.84.
dim(net$generalized.weights[[1]])
## [1] 398 60
max(net$generalized.weights[[1]])
## [1] 57.94401
min(net$generalized.weights[[1]])
## [1] -57.95416
接下来调用gwplot函数可视化泛化权值。其原理:networ$generalized.weights的泛化权值图,四个子图分别展示了30个协变量对diagnosis的响应,如果图中所示的所有泛化值都接近于0,则说明协变量对分类结果影响不大。然后若总体方差大于1,则意味着协变量对分类结果存在非线性影响。
pdf("gwplot.pdf", width = 6, height = 45)
par(mfrow = c(15, 2))
for (i in1:30) {
gwplot(net, selected.covariate = n[i])
}
dev.off()
## png
## 2
测试集测试
基于neuralnet 包得到的模型实现类标号预测,基于一个已经训练好的神经网络和测试数据集test_data生成相关的预测概率矩阵,然后通过找到概率最大的那一列,得到其他可能的类别,根据预测得到的类标号和实际测试数据集的类标号产生分类表,如下:
#### 预测
predict_net_test <- neuralnet::compute(net, test_data)
predict_result <- round(predict_net_test$net.result, digits = 0)
net.prediction = c("B", "M")[apply(predict_result, 1, which.max)]
net.prediction
## [1] "M" "M" "M" "M" "B" "M" "M" "B" "B" "M" "M" "M" "M" "M" "M" "B" "M" "M"
## [19] "B" "B" "M" "M" "B" "B" "M" "B" "M" "B" "B" "B" "B" "M" "B" "B" "M" "B"
## [37] "B" "B" "B" "B" "M" "M" "B" "B" "B" "B" "B" "B" "B" "B" "B" "B" "B" "M"
## [55] "B" "B" "M" "B" "B" "B" "M" "M" "M" "B" "M" "B" "M" "M" "B" "B" "B" "B"
## [73] "B" "M" "B" "B" "M" "M" "M" "M" "M" "M" "M" "M" "B" "B" "B" "M" "M" "B"
## [91] "B" "M" "B" "B" "B" "B" "B" "B" "B" "B" "M" "B" "M" "B" "B" "M" "M" "M"
## [109] "B" "M" "M" "B" "B" "B" "B" "M" "M" "B" "B" "B" "B" "B" "M" "B" "B" "B"
## [127] "B" "B" "B" "B" "B" "B" "B" "B" "B" "M" "B" "M" "M" "M" "B" "B" "B" "B"
## [145] "B" "B" "B" "B" "M" "B" "B" "B" "B" "B" "B" "M" "B" "B" "B" "B" "B" "B"
## [163] "B" "B" "B" "B" "B" "B" "B" "M"
predict.table = table(data$diagnosis[-train_id], net.prediction)
predict.table
## net.prediction
## B M
## B 103 4
## M 8 55
同样也可以通过交叉表格来个预测结果,如下:
library(gmodels)
CrossTable(x = data$diagnosis[-train_id], y = net.prediction, prop.chisq = FALSE)
##
##
## Cell Contents
## |-------------------------|
## | N |
## | N / Row Total |
## | N / Col Total |
## | N / Table Total |
## |-------------------------|
##
##
## Total Observations in Table: 170
##
##
## | net.prediction
## data$diagnosis[-train_id] | B | M | Row Total |
## --------------------------|-----------|-----------|-----------|
## B | 103 | 4 | 107 |
## | 0.963 | 0.037 | 0.629 |
## | 0.928 | 0.068 | |
## | 0.606 | 0.024 | |
## --------------------------|-----------|-----------|-----------|
## M | 8 | 55 | 63 |
## | 0.127 | 0.873 | 0.371 |
## | 0.072 | 0.932 | |
## | 0.047 | 0.324 | |
## --------------------------|-----------|-----------|-----------|
## Column Total | 111 | 59 | 170 |
## | 0.653 | 0.347 | |
## --------------------------|-----------|-----------|-----------|
##
##
绘制ROC曲线
根据测试的预测结果绘制ROC曲线,我们这里使用的是ROSE软件包绘制ROC曲线,如下:
### 绘制ROC曲线
library(ROSE)
roc.curve(net.prediction, data$diagnosis[-train_id], main = "ROC curve of neuralNet",
col = 2, lwd = 2, lty = 2)
## Area under the curve (AUC): 0.930
legend("bottomright", "AUC:0.970", col = 2, bty = "n")
结果解读
我们再回忆一下对乳腺癌数据我们都用过哪些机器学习方法。
基于乳腺癌的数据我们已经做过四种类型的机器学习算法,如下:
K-邻近算法(KNN)准确率为0.9471;
支持向量机(SVM)准确率为 0.9765
分类随机森林RT准确率为0.969
梯度提升准确率为 0.997,准确率嗖的一下就上去了,这模型应该是让老板十万分满意。
这次我们有使用了神经网络准确率为0.970,准确率也算可以,但是比起来发现梯度提升效果更好!
其实这说明做模型时,不应单一只是用一种算法,需要多种算法比较,找到最优的选择!
还有就是注意绘制ROC的方法,在做分类随机森林和神经网络时我们使用的是ROSE软件包,而在做回归随机森林时我们使用的是ROCR,需要注意使用时方法的选择。在这里我们使用InformationValue软件包里面的plotROC.
References:
-
Riedmiller M. (1994) Rprop - Description and Implementation Details. Technical Report. University of Karlsruhe.
-
Riedmiller M. and Braun H. (1993) A direct adaptive method for faster backpropagation learning: The RPROP algorithm. Proceedings of the IEEE International Conference on Neural Networks (ICNN), pages 586-591. San Francisco.
-
Anastasiadis A. et. al. (2005) New globally convergent training scheme based on the resilient propagation algorithm. Neurocomputing 64, pages 253-270.
-
Intrator O. and Intrator N. (1993) Using Neural Nets for Interpretation of Nonlinear Models. Proceedings of the Statistical Computing Section, 244-249 San Francisco: American Statistical Society (eds).