阿里云天池大赛赛题(机器学习)——工业蒸汽量预测(完整代码)
目录
- 赛题背景
- 全代码
-
- 导入包
- 导入数据
- 合并数据
- 删除相关特征
- 数据最大最小归一化
- 画图:探查特征和标签相关信息
- 对特征进行Box-Cox变换,使其满足正态性
-
- 标签数据统计转换后的数据,计算分位数画图展示(基于正态分布)
- 标签数据对数变换数据,使数据更符合正态,并画图展示
- 获取训练和测试数据
- 评分函数
- 获取异常数据,并画图
- 使用删除异常的数据进行模型训练
- 采用网格搜索训练模型
- 岭回归
- Lasso回归
- ElasticNet 回归
- SVR回归
- K近邻
- GBDT 模型
- XGB模型
- 随机森林模型
- 模型预测--多模型Bagging
- Bagging预测
- 模型融合Stacking
-
- 模型融合stacking简单示例
- 工业蒸汽多模型融合stacking
- 模型融合stacking基学习器
- 模型融合stacking预测
- 加载数据
- K折交叉验证
- 训练集和测试集数据
- 使用lr_reg和lgb_reg进行融合预测
赛题背景
火力发电就是燃料燃烧加热水生成蒸汽,蒸汽产生的压力推动汽轮机旋转,进而带动电机旋转,产生电能。其中一系列的能量转化中,影响发电效率的核心是锅炉的燃烧效率,即加热水产生的蒸汽量。而影响锅炉燃烧效率的因素很多,包括锅炉床温、床压、炉膛温度、压力等等。
这个赛题的目标就是给一堆锅炉传感器采集的数据(38个特征变量),然后用训练好的模型预测出蒸汽量。因为预测值为连续型数值变量,且给定的数据都带有标签,故此问题是典型的回归预测问题。
典型的回归预测模型使用的算法包括:线性回归,岭回归,LASSO回归,决策树回归,梯度提升树回归。
全代码
一个典型的机器学习实战算法基本包括 1) 数据处理,2) 特征选取、优化,和 3) 模型选取、验证、优化。 因为 “数据和特征决定了机器学习的上限,而模型和算法知识逼近这个上限而已。” 所以在解决一个机器学习问题时大部分时间都会花在数据处理和特征优化上。
大家最好在jupyter notebook上一段一段地跑下面的代码,加深理解。
机器学习的基本知识可以康康我的其他文章哦 好康的。
导入包
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
plt.rcParams.update({'figure.max_open_warning': 0})
import seaborn as sns
# modelling
import pandas as pd
import numpy as np
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV, RepeatedKFold, cross_val_score,cross_val_predict,KFold
from sklearn.metrics import make_scorer,mean_squared_error
from sklearn.linear_model import LinearRegression, Lasso, Ridge, ElasticNet
from sklearn.svm import LinearSVR, SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor,AdaBoostRegressor
from xgboost import XGBRegressor
from sklearn.preprocessing import PolynomialFeatures,MinMaxScaler,StandardScaler
导入数据
#load_dataset
with open("./zhengqi_train.txt") as fr:
data_train=pd.read_table(fr,sep="\t")
with open("./zhengqi_test.txt") as fr_test:
data_test=pd.read_table(fr_test,sep="\t")
合并数据
#merge train_set and test_set
data_train["oringin"]="train"
data_test["oringin"]="test"
data_all=pd.concat([data_train,data_test],axis=0,ignore_index=True)
删除相关特征
data_all.drop(["V5","V9","V11","V17","V22","V28"],axis=1,inplace=True)
数据最大最小归一化
# normalise numeric columns
cols_numeric=list(data_all.columns)
cols_numeric.remove("oringin")
def scale_minmax(col):
return (col-col.min())/(col.max()-col.min())
scale_cols = [col for col in cols_numeric if col!='target']
data_all[scale_cols] = data_all[scale_cols].apply(scale_minmax,axis=0)
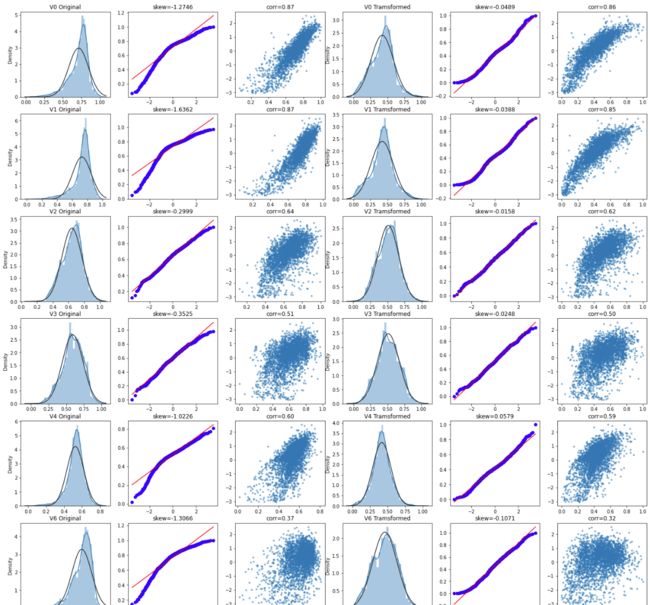
画图:探查特征和标签相关信息
#Check effect of Box-Cox transforms on distributions of continuous variables
fcols = 6
frows = len(cols_numeric)-1
plt.figure(figsize=(4*fcols,4*frows))
i=0
for var in cols_numeric:
if var!='target':
dat = data_all[[var, 'target']].dropna()
i+=1
plt.subplot(frows,fcols,i)
sns.distplot(dat[var] , fit=stats.norm);
plt.title(var+' Original')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(dat[var], plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(dat[var])))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(dat[var], dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(dat[var], dat['target'])[0][1]))
i+=1
plt.subplot(frows,fcols,i)
trans_var, lambda_var = stats.boxcox(dat[var].dropna()+1)
trans_var = scale_minmax(trans_var)
sns.distplot(trans_var , fit=stats.norm);
plt.title(var+' Tramsformed')
plt.xlabel('')
i+=1
plt.subplot(frows,fcols,i)
_=stats.probplot(trans_var, plot=plt)
plt.title('skew='+'{:.4f}'.format(stats.skew(trans_var)))
plt.xlabel('')
plt.ylabel('')
i+=1
plt.subplot(frows,fcols,i)
plt.plot(trans_var, dat['target'],'.',alpha=0.5)
plt.title('corr='+'{:.2f}'.format(np.corrcoef(trans_var,dat['target'])[0][1]))
对特征进行Box-Cox变换,使其满足正态性
Box-Cox变换是Box和Cox在1964年提出的一种广义幂变换方法,是统计建模中常用的一种数据变换,用于连续的响应变量不满足正态分布的情况。Box-Cox变换之后,可以一定程度上减小不可观测的误差和预测变量的相关性。Box-Cox变换的主要特点是引入一个参数,通过数据本身估计该参数进而确定应采取的数据变换形式,Box-Cox变换可以明显地改善数据的正态性、对称性和方差相等性,对许多实际数据都是行之有效的。
cols_transform=data_all.columns[0:-2]
for col in cols_transform:
# transform column
data_all.loc[:,col], _ = stats.boxcox(data_all.loc[:,col]+1)
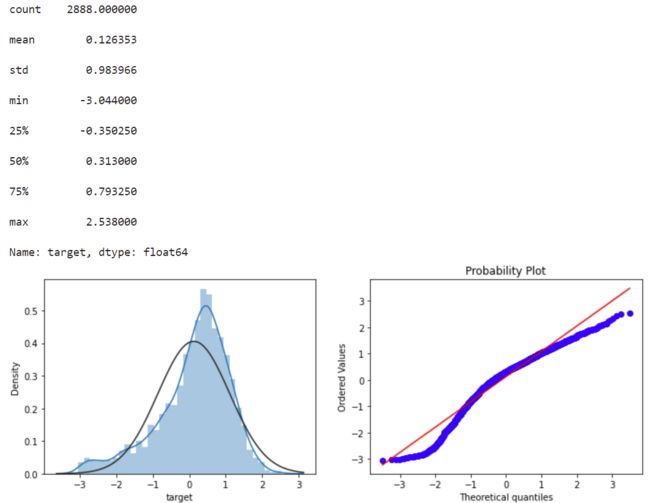
标签数据统计转换后的数据,计算分位数画图展示(基于正态分布)
print(data_all.target.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_all.target.dropna() , fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_all.target.dropna(), plot=plt)
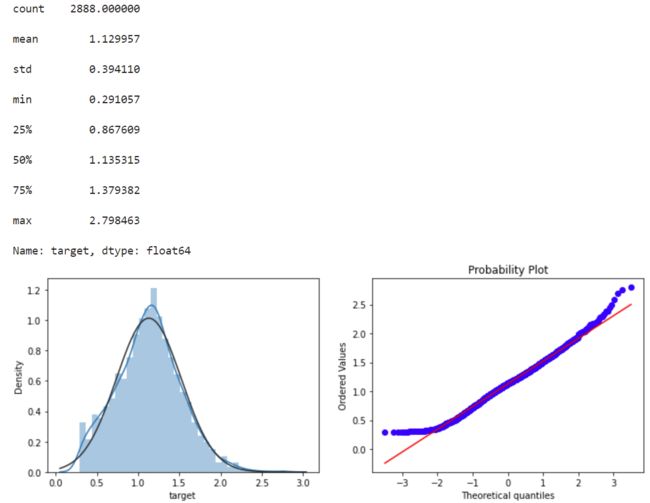
标签数据对数变换数据,使数据更符合正态,并画图展示
#Log Transform SalePrice to improve normality
sp = data_train.target
data_train.target1 =np.power(1.5,sp)
print(data_train.target1.describe())
plt.figure(figsize=(12,4))
plt.subplot(1,2,1)
sns.distplot(data_train.target1.dropna(),fit=stats.norm);
plt.subplot(1,2,2)
_=stats.probplot(data_train.target1.dropna(), plot=plt)
获取训练和测试数据
# function to get training samples
def get_training_data():
# extract training samples
from sklearn.model_selection import train_test_split
df_train = data_all[data_all["oringin"]=="train"]
df_train["label"]=data_train.target1
# split SalePrice and features
y = df_train.target
X = df_train.drop(["oringin","target","label"],axis=1)
X_train,X_valid,y_train,y_valid=train_test_split(X,y,test_size=0.3,random_state=100)
return X_train,X_valid,y_train,y_valid
# extract test data (without SalePrice)
def get_test_data():
df_test = data_all[data_all["oringin"]=="test"].reset_index(drop=True)
return df_test.drop(["oringin","target"],axis=1)
评分函数
from sklearn.metrics import make_scorer
# metric for evaluation
def rmse(y_true, y_pred):
diff = y_pred - y_true
sum_sq = sum(diff**2)
n = len(y_pred)
return np.sqrt(sum_sq/n)
def mse(y_ture,y_pred):
return mean_squared_error(y_ture,y_pred)
# scorer to be used in sklearn model fitting
rmse_scorer = make_scorer(rmse, greater_is_better=False)
mse_scorer = make_scorer(mse, greater_is_better=False)
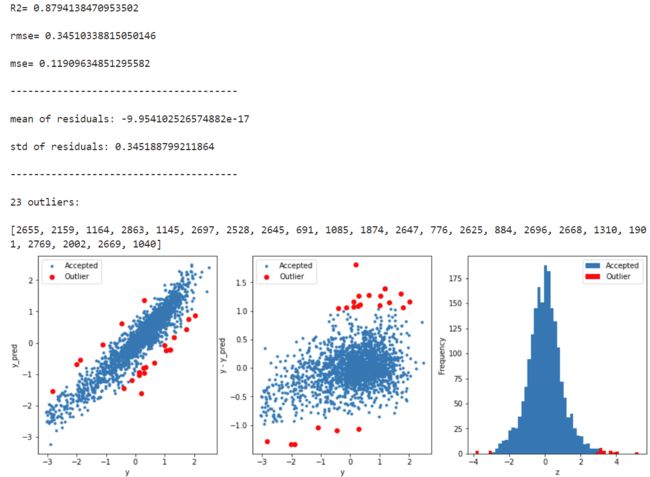
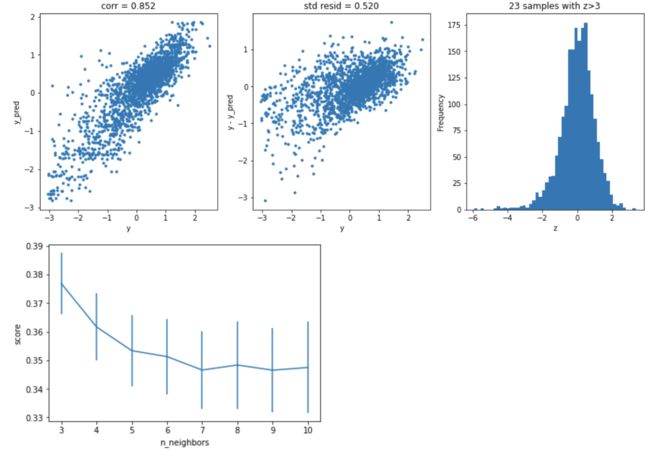
获取异常数据,并画图
# function to detect outliers based on the predictions of a model
def find_outliers(model, X, y, sigma=3):
# predict y values using model
try:
y_pred = pd.Series(model.predict(X), index=y.index)
# if predicting fails, try fitting the model first
except:
model.fit(X,y)
y_pred = pd.Series(model.predict(X), index=y.index)
# calculate residuals between the model prediction and true y values
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
# calculate z statistic, define outliers to be where |z|>sigma
z = (resid - mean_resid)/std_resid
outliers = z[abs(z)>sigma].index
# print and plot the results
print('R2=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print("mse=",mean_squared_error(y,y_pred))
print('---------------------------------------')
print('mean of residuals:',mean_resid)
print('std of residuals:',std_resid)
print('---------------------------------------')
print(len(outliers),'outliers:')
print(outliers.tolist())
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.plot(y.loc[outliers],y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y_pred');
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y - y_pred');
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
z.loc[outliers].plot.hist(color='r',bins=50,ax=ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
plt.savefig('outliers.png')
return outliers
# get training data
from sklearn.linear_model import Ridge
X_train, X_valid,y_train,y_valid = get_training_data()
test=get_test_data()
# find and remove outliers using a Ridge model
outliers = find_outliers(Ridge(), X_train, y_train)
# permanently remove these outliers from the data
#df_train = data_all[data_all["oringin"]=="train"]
#df_train["label"]=data_train.target1
#df_train=df_train.drop(outliers)
X_outliers=X_train.loc[outliers]
y_outliers=y_train.loc[outliers]
X_t=X_train.drop(outliers)
y_t=y_train.drop(outliers)
使用删除异常的数据进行模型训练
def get_trainning_data_omitoutliers():
y1=y_t.copy()
X1=X_t.copy()
return X1,y1
采用网格搜索训练模型
from sklearn.preprocessing import StandardScaler
def train_model(model, param_grid=[], X=[], y=[],
splits=5, repeats=5):
# get unmodified training data, unless data to use already specified
if len(y)==0:
X,y = get_trainning_data_omitoutliers()
#poly_trans=PolynomialFeatures(degree=2)
#X=poly_trans.fit_transform(X)
#X=MinMaxScaler().fit_transform(X)
# create cross-validation method
rkfold = RepeatedKFold(n_splits=splits, n_repeats=repeats)
# perform a grid search if param_grid given
if len(param_grid)>0:
# setup grid search parameters
gsearch = GridSearchCV(model, param_grid, cv=rkfold,
scoring="neg_mean_squared_error",
verbose=1, return_train_score=True)
# search the grid
gsearch.fit(X,y)
# extract best model from the grid
model = gsearch.best_estimator_
best_idx = gsearch.best_index_
# get cv-scores for best model
grid_results = pd.DataFrame(gsearch.cv_results_)
cv_mean = abs(grid_results.loc[best_idx,'mean_test_score'])
cv_std = grid_results.loc[best_idx,'std_test_score']
# no grid search, just cross-val score for given model
else:
grid_results = []
cv_results = cross_val_score(model, X, y, scoring="neg_mean_squared_error", cv=rkfold)
cv_mean = abs(np.mean(cv_results))
cv_std = np.std(cv_results)
# combine mean and std cv-score in to a pandas series
cv_score = pd.Series({'mean':cv_mean,'std':cv_std})
# predict y using the fitted model
y_pred = model.predict(X)
# print stats on model performance
print('----------------------')
print(model)
print('----------------------')
print('score=',model.score(X,y))
print('rmse=',rmse(y, y_pred))
print('mse=',mse(y, y_pred))
print('cross_val: mean=',cv_mean,', std=',cv_std)
# residual plots
y_pred = pd.Series(y_pred,index=y.index)
resid = y - y_pred
mean_resid = resid.mean()
std_resid = resid.std()
z = (resid - mean_resid)/std_resid
n_outliers = sum(abs(z)>3)
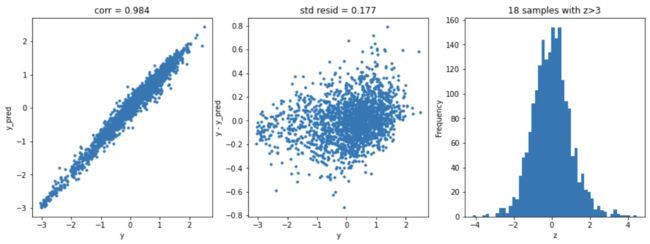
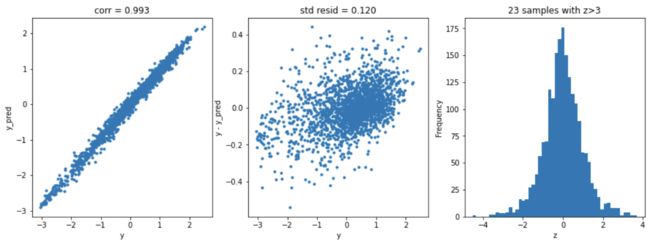
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')
plt.xlabel('y')
plt.ylabel('y_pred');
plt.title('corr = {:.3f}'.format(np.corrcoef(y,y_pred)[0][1]))
ax_132=plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.xlabel('y')
plt.ylabel('y - y_pred');
plt.title('std resid = {:.3f}'.format(std_resid))
ax_133=plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)
plt.xlabel('z')
plt.title('{:.0f} samples with z>3'.format(n_outliers))
return model, cv_score, grid_results
# places to store optimal models and scores
opt_models = dict()
score_models = pd.DataFrame(columns=['mean','std'])
# no. k-fold splits
splits=5
# no. k-fold iterations
repeats=5
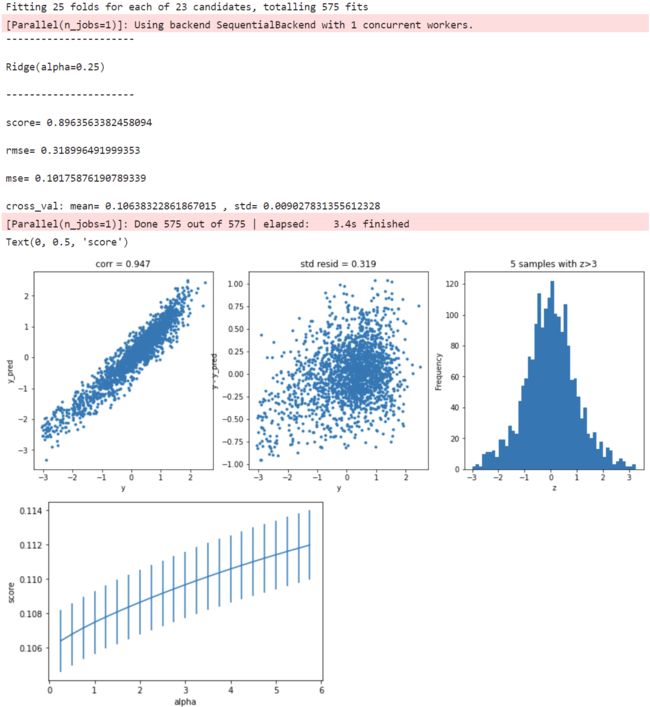
岭回归
model = 'Ridge'
opt_models[model] = Ridge()
alph_range = np.arange(0.25,6,0.25)
param_grid = {'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),
abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
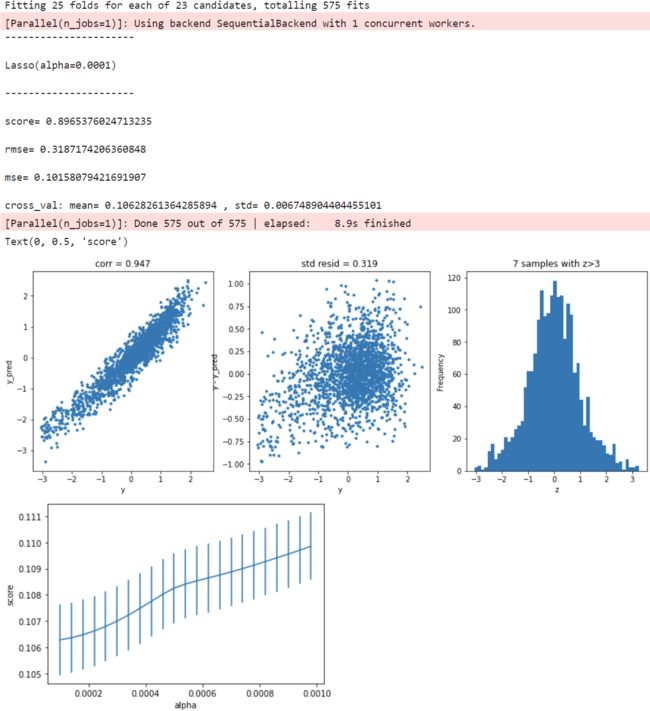
Lasso回归
model = 'Lasso'
opt_models[model] = Lasso()
alph_range = np.arange(1e-4,1e-3,4e-5)
param_grid = {'alpha': alph_range}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range, abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
ElasticNet 回归
model ='ElasticNet'
opt_models[model] = ElasticNet()
param_grid = {'alpha': np.arange(1e-4,1e-3,1e-4),
'l1_ratio': np.arange(0.1,1.0,0.1),
'max_iter':[100000]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
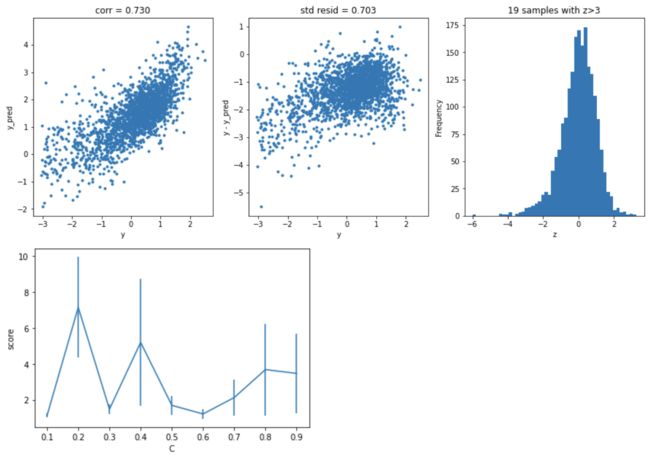
SVR回归
model='LinearSVR'
opt_models[model] = LinearSVR()
crange = np.arange(0.1,1.0,0.1)
param_grid = {'C':crange,
'max_iter':[1000]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(crange, abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('C')
plt.ylabel('score')
K近邻
model = 'KNeighbors'
opt_models[model] = KNeighborsRegressor()
param_grid = {'n_neighbors':np.arange(3,11,1)}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(np.arange(3,11,1), abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*1))
plt.xlabel('n_neighbors')
plt.ylabel('score')
GBDT 模型
model = 'GradientBoosting'
opt_models[model] = GradientBoostingRegressor()
param_grid = {'n_estimators':[150,250,350],
'max_depth':[1,2,3],
'min_samples_split':[5,6,7]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
XGB模型
model = 'XGB'
opt_models[model] = XGBRegressor()
param_grid = {'n_estimators':[100,200,300,400,500],
'max_depth':[1,2,3],
}
opt_models[model], cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=splits, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
随机森林模型
model = 'RandomForest'
opt_models[model] = RandomForestRegressor()
param_grid = {'n_estimators':[100,150,200],
'max_features':[8,12,16,20,24],
'min_samples_split':[2,4,6]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model], param_grid=param_grid,
splits=5, repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
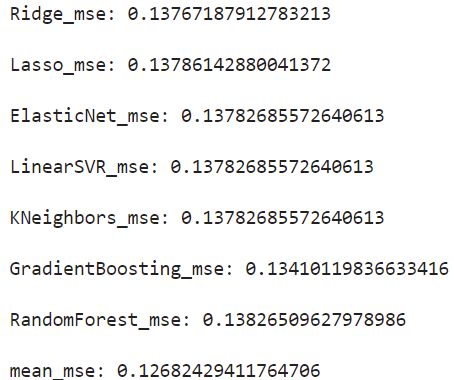
模型预测–多模型Bagging
def model_predict(test_data,test_y=[],stack=False):
#poly_trans=PolynomialFeatures(degree=2)
#test_data1=poly_trans.fit_transform(test_data)
#test_data=MinMaxScaler().fit_transform(test_data)
i=0
y_predict_total=np.zeros((test_data.shape[0],))
for model in opt_models.keys():
if model!="LinearSVR" and model!="KNeighbors":
y_predict=opt_models[model].predict(test_data)
y_predict_total+=y_predict
i+=1
if len(test_y)>0:
print("{}_mse:".format(model),mean_squared_error(y_predict,test_y))
y_predict_mean=np.round(y_predict_total/i,3)
if len(test_y)>0:
print("mean_mse:",mean_squared_error(y_predict_mean,test_y))
else:
y_predict_mean=pd.Series(y_predict_mean)
return y_predict_mean
Bagging预测
model_predict(X_valid,y_valid)
模型融合Stacking
模型融合,即先产生一组个体模型,再用某种策略将它们结合起来,以加强模型效果。
分析表明,随着集成中个体模型数量T增加,集成模型的错误率将呈指数级下降,最终趋于0。通过融合可以达到取长补短的效果,综合个体模型的优势能降低预测误差、优化整体模型的性能。而且个体模型的准确率越高,多样性越大,模型融合的提升效果就越好!
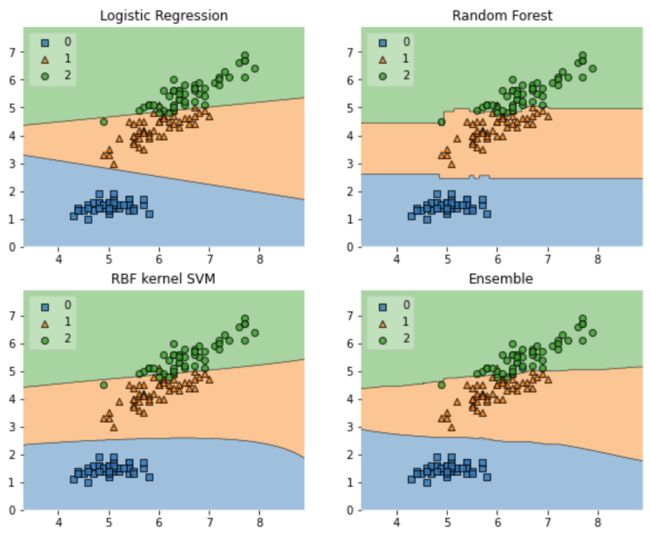
模型融合stacking简单示例
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import itertools
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
##主要使用pip install mlxtend安装mlxtend
from mlxtend.classifier import EnsembleVoteClassifier
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
%matplotlib inline
# Initializing Classifiers
clf1 = LogisticRegression(random_state=0)
clf2 = RandomForestClassifier(random_state=0)
clf3 = SVC(random_state=0, probability=True)
eclf = EnsembleVoteClassifier(clfs=[clf1, clf2, clf3], weights=[2, 1, 1], voting='soft')
# Loading some example data
X, y = iris_data()
X = X[:,[0, 2]]
# Plotting Decision Regions
gs = gridspec.GridSpec(2, 2)
fig = plt.figure(figsize=(10, 8))
for clf, lab, grd in zip([clf1, clf2, clf3, eclf],
['Logistic Regression', 'Random Forest', 'RBF kernel SVM', 'Ensemble'],
itertools.product([0, 1], repeat=2)):
clf.fit(X, y)
ax = plt.subplot(gs[grd[0], grd[1]])
fig = plot_decision_regions(X=X, y=y, clf=clf, legend=2)
plt.title(lab)
plt.show()
工业蒸汽多模型融合stacking
from sklearn.model_selection import train_test_split
import pandas as pd
import numpy as np
from scipy import sparse
import xgboost
import lightgbm
from sklearn.ensemble import RandomForestRegressor,AdaBoostRegressor,GradientBoostingRegressor,ExtraTreesRegressor
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
def stacking_reg(clf,train_x,train_y,test_x,clf_name,kf,label_split=None):
train=np.zeros((train_x.shape[0],1))
test=np.zeros((test_x.shape[0],1))
test_pre=np.empty((folds,test_x.shape[0],1))
cv_scores=[]
for i,(train_index,test_index) in enumerate(kf.split(train_x,label_split)):
tr_x=train_x[train_index]
tr_y=train_y[train_index]
te_x=train_x[test_index]
te_y = train_y[test_index]
if clf_name in ["rf","ada","gb","et","lr","lsvc","knn"]:
clf.fit(tr_x,tr_y)
pre=clf.predict(te_x).reshape(-1,1)
train[test_index]=pre
test_pre[i,:]=clf.predict(test_x).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["xgb"]:
train_matrix = clf.DMatrix(tr_x, label=tr_y, missing=-1)
test_matrix = clf.DMatrix(te_x, label=te_y, missing=-1)
z = clf.DMatrix(test_x, label=te_y, missing=-1)
params = {'booster': 'gbtree',
'eval_metric': 'rmse',
'gamma': 1,
'min_child_weight': 1.5,
'max_depth': 5,
'lambda': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'eta': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12
}
num_round = 10000
early_stopping_rounds = 100
watchlist = [(train_matrix, 'train'),
(test_matrix, 'eval')
]
if test_matrix:
model = clf.train(params, train_matrix, num_boost_round=num_round,evals=watchlist,
early_stopping_rounds=early_stopping_rounds
)
pre= model.predict(test_matrix,ntree_limit=model.best_ntree_limit).reshape(-1,1)
train[test_index]=pre
test_pre[i, :]= model.predict(z, ntree_limit=model.best_ntree_limit).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["lgb"]:
train_matrix = clf.Dataset(tr_x, label=tr_y)
test_matrix = clf.Dataset(te_x, label=te_y)
#z = clf.Dataset(test_x, label=te_y)
#z=test_x
params = {
'boosting_type': 'gbdt',
'objective': 'regression_l2',
'metric': 'mse',
'min_child_weight': 1.5,
'num_leaves': 2**5,
'lambda_l2': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'learning_rate': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12,
'silent': True,
}
num_round = 10000
early_stopping_rounds = 100
if test_matrix:
model = clf.train(params, train_matrix,num_round,valid_sets=test_matrix,
early_stopping_rounds=early_stopping_rounds
)
pre= model.predict(te_x,num_iteration=model.best_iteration).reshape(-1,1)
train[test_index]=pre
test_pre[i, :]= model.predict(test_x, num_iteration=model.best_iteration).reshape(-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
else:
raise IOError("Please add new clf.")
print("%s now score is:"%clf_name,cv_scores)
test[:]=test_pre.mean(axis=0)
print("%s_score_list:"%clf_name,cv_scores)
print("%s_score_mean:"%clf_name,np.mean(cv_scores))
return train.reshape(-1,1),test.reshape(-1,1)
模型融合stacking基学习器
def rf_reg(x_train, y_train, x_valid, kf, label_split=None):
randomforest = RandomForestRegressor(n_estimators=600, max_depth=20, n_jobs=-1, random_state=2017, max_features="auto",verbose=1)
rf_train, rf_test = stacking_reg(randomforest, x_train, y_train, x_valid, "rf", kf, label_split=label_split)
return rf_train, rf_test,"rf_reg"
def ada_reg(x_train, y_train, x_valid, kf, label_split=None):
adaboost = AdaBoostRegressor(n_estimators=30, random_state=2017, learning_rate=0.01)
ada_train, ada_test = stacking_reg(adaboost, x_train, y_train, x_valid, "ada", kf, label_split=label_split)
return ada_train, ada_test,"ada_reg"
def gb_reg(x_train, y_train, x_valid, kf, label_split=None):
gbdt = GradientBoostingRegressor(learning_rate=0.04, n_estimators=100, subsample=0.8, random_state=2017,max_depth=5,verbose=1)
gbdt_train, gbdt_test = stacking_reg(gbdt, x_train, y_train, x_valid, "gb", kf, label_split=label_split)
return gbdt_train, gbdt_test,"gb_reg"
def et_reg(x_train, y_train, x_valid, kf, label_split=None):
extratree = ExtraTreesRegressor(n_estimators=600, max_depth=35, max_features="auto", n_jobs=-1, random_state=2017,verbose=1)
et_train, et_test = stacking_reg(extratree, x_train, y_train, x_valid, "et", kf, label_split=label_split)
return et_train, et_test,"et_reg"
def lr_reg(x_train, y_train, x_valid, kf, label_split=None):
lr_reg=LinearRegression(n_jobs=-1)
lr_train, lr_test = stacking_reg(lr_reg, x_train, y_train, x_valid, "lr", kf, label_split=label_split)
return lr_train, lr_test, "lr_reg"
def xgb_reg(x_train, y_train, x_valid, kf, label_split=None):
xgb_train, xgb_test = stacking_reg(xgboost, x_train, y_train, x_valid, "xgb", kf, label_split=label_split)
return xgb_train, xgb_test,"xgb_reg"
def lgb_reg(x_train, y_train, x_valid, kf, label_split=None):
lgb_train, lgb_test = stacking_reg(lightgbm, x_train, y_train, x_valid, "lgb", kf, label_split=label_split)
return lgb_train, lgb_test,"lgb_reg"
模型融合stacking预测
def stacking_pred(x_train, y_train, x_valid, kf, clf_list, label_split=None, clf_fin="lgb", if_concat_origin=True):
for k, clf_list in enumerate(clf_list):
clf_list = [clf_list]
column_list = []
train_data_list=[]
test_data_list=[]
for clf in clf_list:
train_data,test_data,clf_name=clf(x_train, y_train, x_valid, kf, label_split=label_split)
train_data_list.append(train_data)
test_data_list.append(test_data)
column_list.append("clf_%s" % (clf_name))
train = np.concatenate(train_data_list, axis=1)
test = np.concatenate(test_data_list, axis=1)
if if_concat_origin:
train = np.concatenate([x_train, train], axis=1)
test = np.concatenate([x_valid, test], axis=1)
print(x_train.shape)
print(train.shape)
print(clf_name)
print(clf_name in ["lgb"])
if clf_fin in ["rf","ada","gb","et","lr","lsvc","knn"]:
if clf_fin in ["rf"]:
clf = RandomForestRegressor(n_estimators=600, max_depth=20, n_jobs=-1, random_state=2017, max_features="auto",verbose=1)
elif clf_fin in ["ada"]:
clf = AdaBoostRegressor(n_estimators=30, random_state=2017, learning_rate=0.01)
elif clf_fin in ["gb"]:
clf = GradientBoostingRegressor(learning_rate=0.04, n_estimators=100, subsample=0.8, random_state=2017,max_depth=5,verbose=1)
elif clf_fin in ["et"]:
clf = ExtraTreesRegressor(n_estimators=600, max_depth=35, max_features="auto", n_jobs=-1, random_state=2017,verbose=1)
elif clf_fin in ["lr"]:
clf = LinearRegression(n_jobs=-1)
clf.fit(train, y_train)
pre = clf.predict(test).reshape(-1,1)
return pred
elif clf_fin in ["xgb"]:
clf = xgboost
train_matrix = clf.DMatrix(train, label=y_train, missing=-1)
test_matrix = clf.DMatrix(train, label=y_train, missing=-1)
params = {'booster': 'gbtree',
'eval_metric': 'rmse',
'gamma': 1,
'min_child_weight': 1.5,
'max_depth': 5,
'lambda': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'eta': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12
}
num_round = 10000
early_stopping_rounds = 100
watchlist = [(train_matrix, 'train'),
(test_matrix, 'eval')
]
model = clf.train(params, train_matrix, num_boost_round=num_round,evals=watchlist,
early_stopping_rounds=early_stopping_rounds
)
pre = model.predict(test,ntree_limit=model.best_ntree_limit).reshape(-1,1)
return pre
elif clf_fin in ["lgb"]:
print(clf_name)
clf = lightgbm
train_matrix = clf.Dataset(train, label=y_train)
test_matrix = clf.Dataset(train, label=y_train)
params = {
'boosting_type': 'gbdt',
'objective': 'regression_l2',
'metric': 'mse',
'min_child_weight': 1.5,
'num_leaves': 2**5,
'lambda_l2': 10,
'subsample': 0.7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'learning_rate': 0.03,
'tree_method': 'exact',
'seed': 2017,
'nthread': 12,
'silent': True,
}
num_round = 10000
early_stopping_rounds = 100
model = clf.train(params, train_matrix,num_round,valid_sets=test_matrix,
early_stopping_rounds=early_stopping_rounds
)
print('pred')
pre = model.predict(test,num_iteration=model.best_iteration).reshape(-1,1)
print(pre)
return pre
加载数据
# #load_dataset
with open("./zhengqi_train.txt") as fr:
data_train=pd.read_table(fr,sep="\t")
with open("./zhengqi_test.txt") as fr_test:
data_test=pd.read_table(fr_test,sep="\t")
K折交叉验证
from sklearn.model_selection import StratifiedKFold, KFold
folds = 5
seed = 1
kf = KFold(n_splits=5, shuffle=True, random_state=0)
训练集和测试集数据
x_train = data_train[data_test.columns].values
x_valid = data_test[data_test.columns].values
y_train = data_train['target'].values
使用lr_reg和lgb_reg进行融合预测
clf_list = [lr_reg, lgb_reg]
#clf_list = [lr_reg, rf_reg]
##很容易过拟合
pred = stacking_pred(x_train, y_train, x_valid, kf, clf_list, label_split=None, clf_fin="lgb", if_concat_origin=True)
以上内容和代码全部来自于《阿里云天池大赛赛题解析(机器学习篇)》这本好书,十分推荐大家去阅读原书!