数据挖掘-预测模型汇总
各种预测模型汇总
二、各种预测模型
先总结弄懂了的:
1、naiveBayes(第5课)
#应变量y为email$spam,“~.”表示身下的所有属性都是自变量
#第二个参数我也不知道
#第三个参数为数据源
NBfit<-naiveBayes(as.factor(email$spam)~.,laplace=0,data=email)
#用naiveBayes的结果做预测,第一个参数为用预测函数形成的对象,第二个参数为被预测的自变量的值
#再把预测好的应变量的值存入对象pred2中
pred2<-predict(NBfit,email[,2:19])
#比较预测值与真实值的差别
#形成一个2*2的矩阵,对角线为正确的,其余为预测错误的
table(pred2, email$spam)
2、线性回归(lm函数)-第4课
#createDataPartition函数用于制作训练集,其中y=faithful$waiting指按照该属性分类,p=0.5指将p*100%的值用于训练,list=FALSE不用管,默认为false
inTrain<-createDataPartition(y=faithful$waiting, p=0.5, list=FALSE)
#其中50%用于训练
trainFaith<-faithful[inTrain,]
#剩下50%用于测试
testFaith<-faithful[-inTrain,]
#回归模型函数在这里################################
#同上,y~x,数据源
lm1<-lm(eruptions~waiting, data=trainFaith)
newdata<-data.frame(waiting=80)
predict(lm1, newdata)
注:由于lm函数的预测值是一个置信度为大致0.95左右(可以自己定义)的区间,所以predict共有3个属性结果,所以不可以用它来预测table之类的混淆矩阵
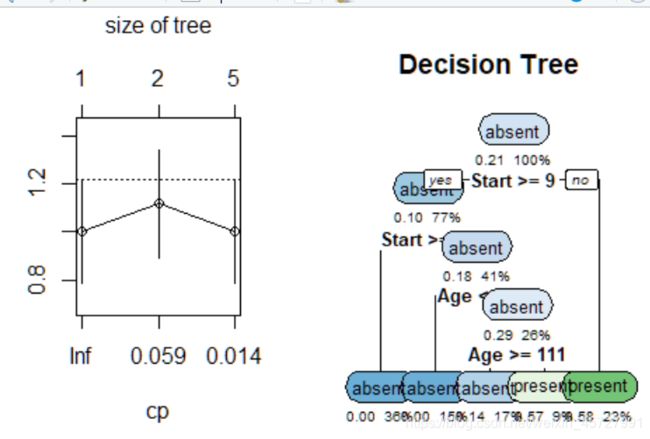
3、决策树:(rpart函数)
# grow the tree
fit <- rpart(Kyphosis ~ Age + Number + Start,
method="class", data=kyphosis)
printcp(fit) # display the results
plotcp(fit) # visualize cross-validation results
summary(fit) # detailed summary of splits
# plot tree
rpart.plot(fit,extra=106, under=TRUE, faclen=0,
cex=0.8, main="Decision Tree")
# prediction
result <- predict(fit,kyphosis[,-1],type="class")
# confusing matrix
table(pred = result, true = kyphosis[,1])
4、剪枝数(是决策树的一种,也是rpart函数,只不过参数中多了一个control = rpart.control(minsplit = 10)约束,其中minsplit表示:试图进行分割时,节点中必须存在的最小观测数。)
# prune the tree
# minsplit: the minimum number of observations that must exist in a node in order for a split to be attempted.
pfit1 <- rpart(Kyphosis ~ Age + Number + Start,
method="class", data=kyphosis,
control = rpart.control(minsplit = 10))
# plot the pruned tree
rpart.plot(pfit1,extra=106, under=TRUE, faclen=0,
cex=0.8, main="Decision Tree")
#prediction
result<-predict(pfit1,kyphosis[,-1],type="class")
# confusing matrix
table(pred = result, true = kyphosis[,1])
5、回归树(也是rpart函数的一种,只不过method=“anova”,只有这点不同)-第7课
fit <- rpart(Mileage~Price + Country + Reliability + Type,
method="anova", data=cu.summary)
6、随机森林(先用n棵树预测,再将这n棵树的结果投票选出最优预测,权重相同,同时进行)-第8课
## Random Forest model
# mtry is the number of variables to try这里的ntree=100就是说要用100棵树先同时预测
fit <- randomForest(y ~ ., data=train, mtry=4, ntree=100)
print(fit) # view results
importance(fit) # importance of each predictor
varImpPlot(fit)
# prediction resutls
RandomTreeresult<-predict(fit,test[,-17],type="class")
# confusing matrix
table(pred = RandomTreeresult, true = test$y)
7、adaboost森林(依次进行不同的预测树,每一次错误时,将权重增大,正确时将权重减小,下一次的值由上一次的值递推得到)-第8课
eg:
adaboost<-boosting(y~., data=train, boos=FALSE, mfinal=20,coeflearn='Breiman')
summary(adaboost)
adaboost$trees
adaboost$weights
adaboost$importance
importanceplot(adaboost)
# error
errorChange<-errorevol(adaboost,train)
plot(errorChange$error,type='l')
# peformance on adTest Data
adpred<-predict(adaboost,test)
table(pred=adpred$class,true=test$y)
# Trees Visulizations: T1,T2, T19, T20
t1<-adaboost$trees[[1]]
t2<-adaboost$trees[[2]]
rpart.plot(t1,under=TRUE, faclen=0,
cex=1, main="Decision Tree 1")
rpart.plot(t2,under=TRUE, faclen=0,
cex=1, main="Decision Tree 2")
2、逻辑回归(Logistic regression)(第5课)
是一种线性的回归
#线性回归分析常用glm函数——逻辑回归
#第一个参数的格式为:y~x,y为应变量,x为自变量,当x为.时,自变量取除了y以外的所有属性
#第二个参数为数据源,第三个参数表示预测集为二元
g_full=glm(spam~ ., data=email, family="binomial")
#接下来进行预测
#confusionMatrix函数的第一个参数表示预测的向量,第二个表示真实的向量
#这个函数的作用是①列出table ②分析精准度等等参数
#由于predict函数预测出来的结果是实数,而这里只需要0/1即可,所以要用ifelse来制约一下
pred<-ifelse(predict(g_full, email[,2:19])>0.5,1,0)
acc <- confusionMatrix(as.factor(pred), as.factor(email$spam))
acc
3、KNN(K Nearest Neighbor Classifiers)(第5课)
维度很高时不适用,运算过程太耗费资源
(想象一个圆圈用于分类不同类型)
# normalize
#标准化
iris_new<-scale(iris_random[,-5],center = TRUE,scale = TRUE)
# data visulization
# 划散点图
ggplot(aes(iris_random$Sepal.Length, iris_random$Petal.Width), data = iris_random)+
geom_point(aes(color= factor(iris_random$Species)))
# constrcut training and testing mannually
#抽出训练和测试数据集
train <- iris_new[1:100,]
test <- iris_new[101:150,]
train_sp <- iris_random[1:100,5]
test_sp <- iris_random[101:150,5]
# knn训练
model <- knn(train= train,test=test,cl= train_sp,k=8)
model
#汇总统计分布
table(factor(model))
#真实和预测的分布
table(test_sp,model)
#choose the right k####
#First Attempt to Determine Right K ####
#存储准确率
iris_acc<-numeric() #Holding variable
#k从1-50取值,看哪一个准确率高
for(i in 1:50){
#Apply knn with k = i
predict<-knn(train= train,test=test,cl= train_sp,k=i)
iris_acc<-c(iris_acc,
mean(predict==iris_random[101:150,5]))
}
#Plot k= 1 through 50
#画图,准确率和k值的关系
plot(1-iris_acc,type="l",ylab="Error Rate",
xlab="K",main="Error Rate for Iris With Varying K")
# 从图像中看出k= 12 时最优
# optimal k =12
model <- knn(train= train,test=test,cl= train_sp,k=12)
table(test_sp,model)
4、perceptron(是一种线性分类器)
##iris DATA
#鸢尾花的那些数据,实验方法和上面的例子相同
data(iris)
pairs(iris[1:4], main = "Anderson's Iris Data -- 3 species",
pch = 21, bg = c("red", "green3", "blue")[unclass(iris$Species)])
# select the Sepal.Width versus Petal.Width scatter plot
x <- cbind(iris$Sepal.Width,iris$Petal.Width)
# label setosa as positive and the rest as negative
Y <- ifelse(iris$Species == "setosa", +1, -1)
# # plot all the points
plot(x,cex=0.5)
# use plus sign for setosa points
points(subset(x,Y==1),col="black",pch="+",cex=2)
# use minus sign for the rest
points(subset(x,Y==-1),col="red",pch="-",cex=2)
p <- perceptron(x,Y)
plot(x,cex=0.2)
points(subset(x,Y==1),col="black",pch="+",cex=2)
points(subset(x,Y==-1),col="red",pch="-",cex=2)
# compute intercept on y axis of separator
# from w and b
intercept <- - p$b / p$w[[2]]
# compute slope of separator from w
slope <- - p$w[[1]] /p$ w[[2]]
# draw separating boundary
abline(intercept,slope,col="red")
5、SVM(Support vector machines)
#subset 1: setosa and virginica
#把其中两种花区分出来(易区分)
iris.part1 = subset(iris, Species != 'versicolor')
pairs(iris.part1[,1:4], col=iris.part1$Species)
iris.part1$Species = factor(iris.part1$Species)
#subset 2: versicolor and virginica
#把另外两种花区分出来(不易区分)
iris.part2 = subset(iris, Species != 'setosa')
pairs(iris.part2[,1:4], col=iris.part2$Species)
iris.part2$Species = factor(iris.part2$Species)
iris.part1 = iris.part1[, c(1,2,5)]
iris.part2 = iris.part2[, c(1,2,5)]
# linear
#线性可分的情况
plot(iris.part1$Sepal.Length,iris.part1$Sepal.Width,col=iris.part1$Species)
fit1 = svm(Species ~ ., data=iris.part1, type='C-classification')
plot(fit1, iris.part1)
fit1.pred<-predict(fit1,iris.part1[,-3])
table(pred = fit1.pred, true = iris.part1[,3])
# linear
#部分线性不可分的情况
fit2 = svm(Species ~ ., data=iris.part2, type='C-classification', kernel='linear')
plot(fit2, iris.part2)
fit2.pred<-predict(fit2,iris.part2[,-3])
table(pred = fit2.pred, true = iris.part2[,3])
# Non-linear
#线性不可分
plot(iris.part2$Sepal.Length,iris.part2$Sepal.Width,col=iris.part2$Species)
fit3 = svm(Species ~ ., data=iris.part2, type='C-classification', kernel='radial')
plot(fit3, iris.part2)
fit3.pred<-predict(fit3,iris.part2[,-3])
table(pred = fit3.pred, true = iris.part2[,3])
# multiple-class
#多类别的判定
svm_model <- svm(Species ~ ., data=iris)
summary(svm_model)
x <- subset(iris, select=-Species)
y <- iris$Species
fit4.pred<-predict(svm_model,x)
table(pred = fit4.pred, true = y)
plot(svm_model, iris, Petal.Width ~ Petal.Length,
slice = list(Sepal.Width = 3, Sepal.Length = 4))
7、拉索回归和ridge回归-第4课(没看懂,mark)
预测方式、方法
1、将二元的y值预测强制转换
##因为predict函数预测出来的值是实数,而结果值只有1、0,这样一来就
##需要ifelse函数用于制约y值了,当大于0.5时y值为1,否则为0
pred<-ifelse(predict(g_full, email[,2:19])>0.5,1,0)
2、混淆矩阵
①table
该函数的两个参数分别为预测的所有y值,和真实的所有y值
table(pred2, email$spam)
②
confusionMatrix函数,参数和table一致,只是需要将格式更改为factor才可行
acc <- confusionMatrix(as.factor(pred2), as.factor(email$spam))
acc