BioCreative比赛:Track2:PubMed文章中的NLM-CHEM全文化学识别(NER)、实体链接(EL)和标引(indexing)
一、任务介绍
命名实体识别是许多复杂知识提取任务的重要组成部分。识别相关生物医学实体的错误是准确检索、分类和进一步理解文本语义(例如关系提取)的关键障碍。化学实体出现在整个生物医学研究文献中,是 PubMed 中最常搜索的实体类型之一。准确自动识别期刊出版物中提到的化学物质有可能转化为许多下游 NLP 任务和生物医学领域的改进
MeSH identifer:MeSH (Medical Subject Headings) is the NLM controlled vocabulary thesaurus used for indexing articles for PubMed. 用于为PubMed的文章建立索引。
任务1:Chemical Identification in full text: predicting all chemicals mentioned in recently published full-text articles, both span (i.e. named entity recognition) and normalization (i.e. entity linking) using MeSH. 命名实体识别+实体链接
任务2:Chemical Indexing prediction task: predicting which chemicals mentioned in recently published full-text articles should be indexed, i.e. appear in the listing of MeSH terms for the document. 二分类预测问题
本方法基于nlp数据处理工具spacy和BioBERT模型,并对biobert模型进行改进,同时完成NER、实体链接和indexing三个任务
二、数据集
数据集来自NLM-Chem语料库,包括150篇发表在PubMed Central Open Access上的全文期刊文章。
有两种类型的注释:
- 化学实体注释,用于化学命名识别任务(NER)
两种格式
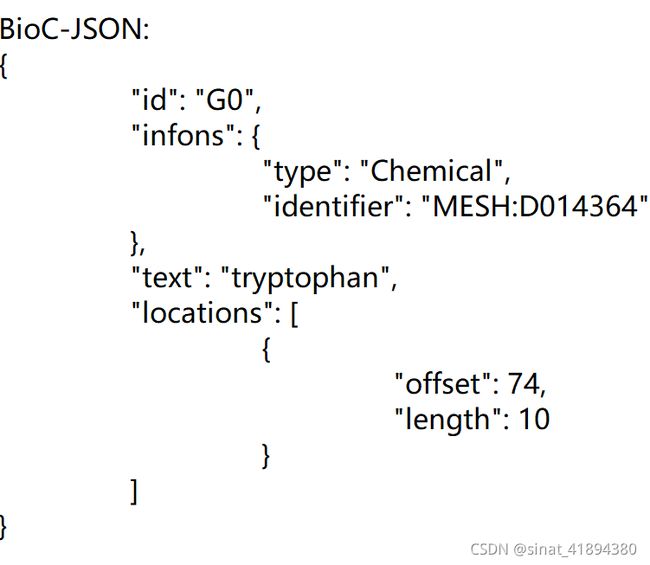
注释类型、对应mesh标识符、位置信息、实体信息
2.网格索引注释,用于化学索引任务
两种格式:
annotation type:’MeSH_Indexing_Chemical’
an annotation identifier
the MeSH identifier
the MeSH terminology entry term
三、实验过程
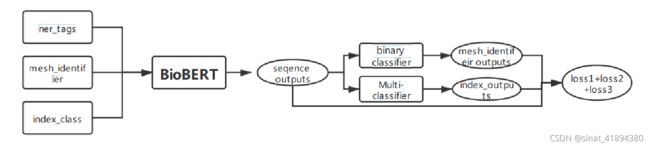
思路:在原有BioBert模型上进行修改,增加实体链接任务和是否被索引预测的loss,三个子任务loss相加统一优化输出预测结果
1、数据集转换
spacy是Python里面的一个工业级别的nlp工具,足见其在自然语言处理方面的优势,所以我们有必要去了解,学习它。Spacy的功能包括词性标注,句法分析,命名实体识别,词向量,与深度学习无缝对接,以及它支持三十多种语言等等
使用spacy需要在环境中安装预训练模型
pip install --user spacy
python -m spacy download en_core_sci_smimport spacy
nlp = spacy.load("en_core_sci_sm")第一步将bioc形式的数据集转换为huggingface中NER模型输入的格式
主要代码:
def write_bert_ner_file(total_sentences, filename):
cnt = 0
elements = []
for sentence in total_sentences:
ner_tags = []
tokens = []
spans = []
for i, ann in enumerate(sentence.annotations):
tokens.append(ann.text)
ner_tags.append(ann.infons.get('NE_label', "O"))
spans.append((ann.total_span.offset, ann.total_span.end))
element = {"id": len(elements), "document_id": sentence.infons["document_id"], "ner_tags": ner_tags, "tokens": tokens, "spans": spans}
elements.append(element)
cnt += 1
with open(filename, 'w') as file:
for element in elements:
file.write(json.dumps(element) + "\n")
return len(elements)
def convert_bioc_to_json(srcs, dest, entity_type = None):
total_sentences = []
for src in srcs:
reader = bioc.BioCXMLDocumentReader(src)
for document in reader:
print("Processing document " + str(document.id) + ", number of sentences = " + str(len(total_sentences)))
for passage in document.passages:
text = passage.text
sentences = tokenize_text(text, document.id, offset=passage.offset)
total_sentences.extend(sentences)
for ann in passage.annotations:
if entity_type is None or ann.infons['type'] == entity_type:
anns = _find_toks(sentences, ann.total_span.offset, ann.total_span.end)
if len(anns) == 0:
logging.debug('%s: Cannot find %s', document.id, ann)
print_ner_debug(sentences, ann.total_span.offset, ann.total_span.end)
continue
has_first = False
for ann in anns:
if not has_first:
ann.infons['NE_label'] = "B-" + entity_type
has_first = True
else:
ann.infons['NE_label'] = "I-" + entity_type
cnt = write_bert_ner_file(total_sentences, dest)
logging.debug("Number of mentions: %s", cnt)
return cnt添加mesh和index的输入信息
entity_type = 'Chemical'
for document in reader:
print("Processing document " + str(document.id) + ", number of sentences = " + str(len(total_sentences)))
for passage in document.passages:
text = passage.text
#display(text) #输出文本
sentences = tokenize_text(text, document.id, offset=passage.offset)
#display(sentences)#分句
#total_sentences.extend(sentences)
total_sentences.extend(sentences)
for ann in passage.annotations:
#meshs=meshs.append(ann.infons['identifier'])
#display(ann.infons['identifier'] )
ann.infons['mesh']=ann.infons['identifier']
if entity_type is None or ann.infons['type'] == entity_type:
anns = _find_toks(sentences, ann.total_span.offset, ann.total_span.end)
if len(anns) == 0:
logging.debug('%s: Cannot find %s', document.id, ann)
print_ner_debug(sentences, ann.total_span.offset, ann.total_span.end)
continue
has_first = False
# print(anns)
#print(ann)
for annn in anns:
mesh_count+=1
#print(annn)
if not has_first:
annn.infons['NE_label'] = "B-" + entity_type
annn.infons['mesh']=ann.infons['identifier']
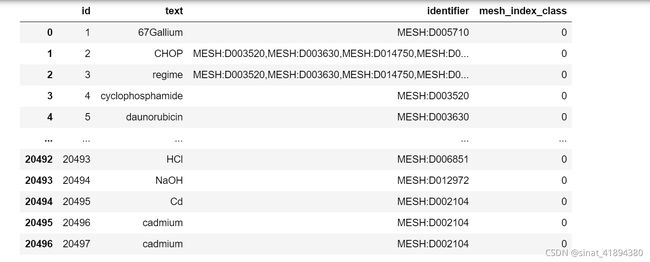
test_dict['id'].append(mesh_count)
test_dict['text'].append(annn.text)
test_dict['identifier'].append(annn.infons['mesh'])
has_first = True
else:
annn.infons['NE_label'] = "I-" + entity_type
annn.infons['mesh']=ann.infons['identifier']
test_dict['id'].append(mesh_count)
test_dict['text'].append(annn.text)
test_dict['identifier'].append(annn.infons['mesh'])
#ann.infons['mesh']=ann.infons['identifier']for document in reader:
print("Processing document " + str(document.id) + ", number of sentences = " + str(len(total_sentences)))
for passage in document.passages:
for ann in passage.annotations:
text=ann.text
#print(type(text))
#print(ann)
#print(ann.infons['type'])
types=ann.infons['type']
if types=='MeSH_Indexing_Chemical':
#print(ann.infons["identifier"])
index_mesh.append(ann.infons["identifier"])
index_entry=ann.infons["entry_term"]
#print(test_dict['identifier'])
i=0
for identifier in test_dict2['identifier']:
if identifier in index_mesh:
#print(identifier)
#print(1)
test_dict2['mesh_index_class'][i]=1
i+=1以json形式存入后
2、数据集处理
数据分析代码:获取label_to_id,id_to_label代码
#获取ner label list
if isinstance(features[label_column_name].feature, ClassLabel):
label_list = features[label_column_name].feature.names
# No need to convert the labels since they are already ints.
label_to_id = {i: i for i in range(len(label_list))}
else:
label_list = get_label_list(datasets["train"][label_column_name])
label_to_id = {l: i for i, l in enumerate(label_list)}
num_labels = len(label_list)
print("LABELS: {}, len: {}".format(label_list, num_labels))
#获取mesh label list
if isinstance(features[label_mesh_column_name].feature, ClassLabel):
label_mesh_list = features[label_mesh_column_name].feature.names
# No need to convert the labels since they are already ints.
label_mesh_to_id = {i: i for i in range(len(label_mesh_list))}
else:
label_mesh_list = get_label_list(datasets["train"][label_mesh_column_name])
label_mesh_to_id = {l: i for i, l in enumerate(label_mesh_list)}
num_mesh_labels = len(label_mesh_list)
print("mesh_LABELS: {}, len: {}".format(label_mesh_list, num_mesh_labels))
#获取index_class label list
if isinstance(features[label_01_column_name].feature, ClassLabel):
label_01_list = features[label_01_column_name].feature.names
# No need to convert the labels since they are already ints.
label_01_to_id = {i: i for i in range(len(label_01_list))}
else:
label_01_list = get_label_list(datasets["train"][label_01_column_name])
label_01_to_id = {l: i for i, l in enumerate(label_01_list)}
index_num_labels = len(label_01_list)
print("index_LABELS: {}, len: {}".format(label_01_list, index_num_labels))
id_to_label = dict(zip(label_to_id.values(), label_to_id.keys()))
id_mesh_to_label = dict(zip(label_mesh_to_id.values(), label_mesh_to_id.keys()))
id_01_to_label = dict(zip(label_01_to_id.values(), label_01_to_id.keys())) #ner labels
labels = []
for i, label in enumerate(examples[label_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels.append(label_ids)
#mesh labels
labels_mesh=[]
for i, label in enumerate(examples[label_mesh_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_mesh_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_mesh_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels_mesh.append(label_ids)
#index labels
labels_01=[]
for i, label in enumerate(examples[label_01_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_01_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_01_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels_01.append(label_ids)编码ner、mesh和index三个任务的labels:
#ner labels
labels = []
for i, label in enumerate(examples[label_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels.append(label_ids)
#mesh labels
labels_mesh=[]
for i, label in enumerate(examples[label_mesh_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_mesh_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_mesh_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels_mesh.append(label_ids)
#index labels
labels_01=[]
for i, label in enumerate(examples[label_01_column_name]):
word_ids = tokenized_inputs.word_ids(batch_index=i)
previous_word_idx = None
label_ids = []
for word_idx in word_ids:
# Special tokens have a word id that is None. We set the label to -100 so they are automatically
# ignored in the loss function.
if word_idx is None:
label_ids.append(-100)
# We set the label for the first token of each word.
elif word_idx != previous_word_idx:
label_ids.append(label_01_to_id[label[word_idx]])
# For the other tokens in a word, we set the label to either the current label or -100, depending on
# the label_all_tokens flag.
else:
label_ids.append(label_01_to_id[label[word_idx]] if data_args.label_all_tokens else -100)
previous_word_idx = word_idx
labels_01.append(label_ids)传入tokenize数据集:
tokenized_inputs["labels_mesh"] = labels_mesh
tokenized_inputs["labels_01"] = labels_01
tokenized_inputs["labels"] = labels3、修改BioBERT模型代码:
主要修改/home1/zhaiwq/anaconda3/envs/biobert/lib/python3.9/site-packages/transformers/models/bert/modeling_bert.py和/home1/zhaiwq/anaconda3/envs/biobert/lib/python3.9/site-packages/transformers/trainer.py
模型修改:class BertForTokenClassification(BertPreTrainedModel):函数中增加两个分类器,一个多分类解决mesh identifer链接问题,一个二分类器解决indexing问题
self.net1 = nn.Sequential(
nn.Linear(768, 512),
nn.ReLU(),
nn.Linear(512, 768),
nn.ReLU(),
nn.Linear(768, 1411),
nn.Softmax(dim=None)
)
#index_class模型
self.net2 = nn.Sequential(
nn.Linear(768, 512),
nn.ReLU(),
nn.Linear(512, 100),
nn.ReLU(),
nn.Linear(100, 2),
nn.Sigmoid()
)sequence_output = outputs[0]
sequence_output = self.dropout(sequence_output)
logits1 = self.classifier(sequence_output)
outputs1=logits1
logits2 = self.net1(sequence_output)
logits3 = self.net2(sequence_output)
loss = None
labels1 = logits1.argmax(2)
labels2 = logits2.argmax(2)
labels3 = logits3.argmax(2)
nlabels1, nlabels2, nlabels3 = [], [], []
for i in range(labels.shape[0]):
nlab1, nlab2, nlab3 = [], [], []
for l, l1, l2, l3 in zip(labels[i], labels1[i], labels2[i], labels3[i]):
if l == -100: continue
nlab1.append(l1.item())
if l1 == 0:
nlab2.append(l2.item())
nlab3.append(l3.item())
else:
nlab2.append(-1)
nlab3.append(-1)
nlabels1.append(nlab1)
nlabels2.append(nlab2)
nlabels3.append(nlab3)
return nlabels1, nlabels2, nlabels3, losstrain.py中调用模型
loss, outputs1, outputs2, outputs3 = self.compute_loss(model, inputs, return_outputs=True)
# ipdb.set_trace()
return outputs1, outputs2, outputs3
loss = loss.mean().detach()
if isinstance(outputs, dict):
logits = tuple(v for k, v in outputs.items() if k not in ignore_keys + ["loss"])
else:
logits = outputs预测:
all_labels1, all_labels2, all_labels3 = [], [], []
# Main evaluation loop
for step, inputs in enumerate(dataloader):
#if step > 10: continue
# Update the observed num examples
observed_batch_size = find_batch_size(inputs)
if observed_batch_size is not None:
observed_num_examples += observed_batch_size
# Prediction step
#ipdb.set_trace()
# loss, logits, labels = self.prediction_step(model, inputs, prediction_loss_only, ignore_keys=ignore_keys)
labs1, labs2, labs3 = self.prediction_step(model, inputs, prediction_loss_only, ignore_keys=ignore_keys)
all_labels1.extend(labs1)
all_labels2.extend(labs2)
all_labels3.extend(labs3)最后结果:
任务1
任务2: