fsbrain的学习笔记
之前分析皮层数据,基于freesurferformats构建了一套自己的脚本,后来偶然间发现了fsbrain,把这些都封装好了,并且还包括可视化。因此,就抽空来学习学习。
1
subjects_dir = fsbrain::get_optional_data_filepath("subjects_dir");
subjects_list = c("subject1", "subject2");
subject_id = 'subject1'; # for function which use one subject only
subjects_dir的截图:
 |
 |
|---|
可以看出,subject文件夹是经freesurfer处理后的格式化输出。
2
# Read from a simple ASCII subjects file (one subject per line, typically no header):
sjl = read.md.subjects("~/data/study1/subjects.txt", header = FALSE);
# Extract the subject identifiers from a FreeSurfer Group Descriptor file, used for the GLM:
sjl = read.md.subjects.from.fsgd("~/data/study1/subjects.fsgd");
# Use the subject identifier column from a demographics file, i.e., a CSV file containing demographics and other covariates (typically age, gender, site, IQ):
demographics = read.md.demographics("~/data/study1/demogr.csv", header = TRUE);
sjl = demographics$participant_id; # or whatever your subject identifier column is called.
这个就是介绍了几种读取subjectID的方法,有一般的txt格式,有freesurfer的fsgd格式,有csv格式。
3
groupdata_nat = group.morph.native(subjects_dir, subjects_list, "thickness", "lh");
group.morph.native函数用法:
group.morph.native( subjects_dir, subjects_list, measure, hemi, format = “curv”, cortex_only = FALSE )
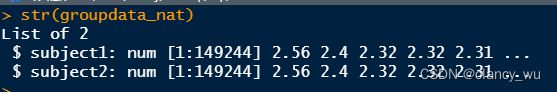
这个函数就很方便了,能循环读取文件夹下所有被试。
length(groupdata_nat[[subject_id]])
> [1] 149244
mean(groupdata_nat[[subject_id]])
> [1] 2.437466
4
groupdata_std = group.morph.standard(subjects_dir, subjects_list, "thickness", "lh", fwhm="10");
这个就是平滑,使用平滑核。
5
# Load the full hemisphere data, including media wall:
fulldata = group.morph.native(subjects_dir, subjects_list, "thickness", "lh");
mean(fulldata$subject1);
# This time, restrict data to the cerebral cortex (the medial wall vertices will get NA values):
cortexdata = group.morph.native(subjects_dir, subjects_list, "thickness", "lh", cortex_only=TRUE);
mean(cortexdata$subject1, na.rm=TRUE);

cortex_only就是将lh.cortex.label传递到函数内,使得内壁等非皮质区域的数值变为0。但是,前提是需要label文件下有cortex.label文件,一般freesurfer都会自动生成。
6
# zipped all the group subjects into one file in the 4th- dimision.
write.group.morph.standard.sf('~/mystudy/group_thickness_lh_fwhm10.mgz', groupdata_std);
#groupdata_std = group.morph.standard(subjects_dir, subjects_list, "thickness", "lh", fwhm="10"); # slow
groupdata_std = group.morph.standard.sf('~/mystudy/group_thickness_lh_fwhm10.mgz'); # fast
这个就是为了更快的处理数据,之前groupdata是list格式,现在把他们全部压缩起来,形成数组,这样操作起来更快,变成了freesurfer的mgz格式。
7
grouplabels = group.label(subjects_dir, subjects_list, "cortex.label", hemi='lh');
从文件夹中加载label文件。
surface = 'white';
hemi = 'both';
atlas = 'aparc';
region = 'bankssts';
# Create a mask from a region of an annotation:
lh_annot = subject.annot(subjects_dir, subject_id, 'lh', atlas);
rh_annot = subject.annot(subjects_dir, subject_id, 'rh', atlas);
lh_label = label.from.annotdata(lh_annot, region);
rh_label = label.from.annotdata(rh_annot, region);
lh_mask = mask.from.labeldata.for.hemi(lh_label, length(lh_annot$vertices));
rh_mask = mask.from.labeldata.for.hemi(rh_label, length(rh_annot$vertices));
# Edit the mask: add the vertices from another region to it:
region2 = 'medialorbitofrontal';
lh_label2 = label.from.annotdata(lh_annot, region2);
rh_label2 = label.from.annotdata(rh_annot, region2);
lh_mask2 = mask.from.labeldata.for.hemi(lh_label2, length(lh_annot$vertices),
existing_mask = lh_mask);
rh_mask2 = mask.from.labeldata.for.hemi(rh_label2, length(rh_annot$vertices),
existing_mask = rh_mask);
这个就是加载ROI了。
groupannot = group.annot(subjects_dir, subjects_list, 'lh', 'aparc');
cat(sprintf("The left hemi of subject2 has %d vertices, and vertex 10 is in region '%s'.\n", length(groupannot$subject2$vertices), groupannot$subject2$label_names[10]));
# output: The left hemi of subject2 has 149244 vertices, and vertex 10 is in region 'lateraloccipital'.
这个就是加载ROI名字了。
8
mean_thickness_lh_native = group.morph.agg.native(subjects_dir, subjects_list, "thickness", "lh", agg_fun=mean);
mean_thickness_lh_native;
# output:
# subject_id hemi measure_name measure_value
#1 subject1 lh thickness 2.437466
#2 subject2 lh thickness 2.437466
这个就是高级用法,agg:aggregate,可以添加一些参数直接对他们运用,类似于lapply函数。
mean_thickness_lh_std = group.morph.agg.standard(subjects_dir, subjects_list, "thickness", "lh", fwhm="10", agg_fun=mean);
mean_thickness_lh_std;
# output:
subject_id hemi measure_name measure_value
1 subject1 lh thickness 2.32443
2 subject2 lh thickness 2.32443
agg_nat = group.multimorph.agg.native(subjects_dir, subjects_list, c("thickness", "area"), c("lh", "rh"), agg_fun = mean);
head(agg_nat);
# output:
# subject_id hemi measure_name measure_value
#1 subject1 lh thickness 2.4374657
#2 subject2 lh thickness 2.4374657
#3 subject1 lh area 0.6690556
#4 subject2 lh area 0.6690556
#5 subject1 rh thickness 2.4143047
#6 subject2 rh thickness 2.4143047
这个就是在原始空间进行一些数据的汇总
data_std = group.multimorph.agg.standard(subjects_dir, subjects_list, c("thickness", "area"), c("lh", "rh"), fwhm='10', agg_fun = mean);
head(data_std);
# output:
# subject_id hemi measure_name measure_value
#1 subject1 lh thickness 2.3244303
#2 subject2 lh thickness 2.3244303
这个是在标准空间进行一些数据的汇总
data_std_cortex = group.multimorph.agg.standard(subjects_dir, subjects_list, c("thickness", "area"), c("lh", "rh"), fwhm='10', agg_fun = mean, cortex_only=TRUE, agg_fun_extra_params=list("na.rm"=TRUE));
head(data_std_cortex);
这个是屏蔽了内壁以后的数据统计,但是注意的是,na.rm一定是需要的。
ROI查看
atlas = 'aparc'; # or 'aparc.a2009s', or 'aparc.DKTatlas'.
measure = 'thickness';
region_means_native = group.agg.atlas.native(subjects_dir, subjects_list, measure, "lh", atlas, agg_fun = mean);
head(region_means_native[,1:6]);
# output:
# subject bankssts caudalanteriorcingulate caudalmiddlefrontal cuneus entorhinal
#subject1 subject1 2.485596 2.70373 2.591197 1.986978 3.702725
#subject2 subject2 2.485596 2.70373 2.591197 1.986978 3.702725
这个是在原始空间查看ROI
region_means_std = group.agg.atlas.standard(subjects_dir, subjects_list, measure, "lh", atlas, fwhm = '10', agg_fun = mean);
head(region_means_std[1:5]);
# output:
# subject bankssts caudalanteriorcingulate caudalmiddlefrontal cuneus
#subject1 subject1 2.583408 2.780666 2.594696 2.018783
#subject2 subject2 2.583408 2.780666 2.594696 2.018783
这个是在标准空间查看ROI
地图集
FreeSurfer 根据您的受试者的三个不同的脑图谱计算脑表面分区:Desikan-Killiany 图集(‘aparc’ 文件,Desikan等人,2006),Destrieux 图集(‘aparc.a2009s’ 文件,Destrieux等al. , 2010) 和 DKT 或 Mindboggle 40 地图集(‘aparc.DKTatlas40’ 文件)
surface = 'white';
hemi = 'both';
atlas = 'aparc'; # one of 'aparc', 'aparc.a2009s', or 'aparc.DKTatlas40'.
region = 'bankssts';
# Create a label from a region of an annotation or atlas:
lh_annot = subject.annot(subjects_dir, subject_id, 'lh', atlas);
rh_annot = subject.annot(subjects_dir, subject_id, 'rh', atlas);
lh_label = label.from.annotdata(lh_annot, region);
rh_label = label.from.annotdata(rh_annot, region);
结果映射
hemi = "lh" # 'lh' or 'rh'
atlas = "aparc" # an atlas, e.g., 'aparc', 'aparc.a2009s', 'aparc.DKTatlas'
# Some directory where we can find fsaverage. This can be omitted if FREESURFER_HOME or SUBJECTS_DIR is set, the function will find fsaverage in there by default. Also see the function download_fsaverage().
template_subjects_dir = "~/software/freesurfer/subjects";
region_value_list = list("bankssts"=0.9, "precuneus"=0.7, "postcentral"=0.8, "lingual"=0.6);
ret = fsbrain::write.region.values.fsaverage(hemi, atlas, region_value_list, output_file="/tmp/lh_spread.mgz", template_subjects_dir=template_subjects_dir, show_freeview_tip=TRUE);
可视化
vis.subject.annot(subjects_dir, 'subject1', 'aparc', 'both', views=c('si'));
surface = 'white';
hemi = 'both';
label = 'cortex.label';
vis.subject.label(subjects_dir, subject_id, label, hemi);
可视化ROI
atlas = 'aparc';
template_subject = 'fsaverage';
# Some directory where we can find the template_subject. This can be omitted if FREESURFER_HOME or SUBJECTS_DIR is set and the template subject is in one of them. In that case, the function will find fsaverage in there by default. Also see the function download_fsaverage().
template_subjects_dir = "~/software/freesurfer/subjects"; # adapt to your machine
# For the left hemi, we manually set data values for some regions.
lh_region_value_list = list("bankssts"=0.9, "precuneus"=0.7, "postcentral"=0.8, "lingual"=0.6);
# For the right hemisphere, we do something a little bit more complex: first get all atlas region names:
atlas_region_names = get.atlas.region.names(atlas, template_subjects_dir=template_subjects_dir, template_subject=template_subject);
# As mentioned above, if you have fsaverage in your SUBJECTS_DIR or FREESURFER_HOME is set, you could replace the last line with:
#atlas_region_names = get.atlas.region.names(atlas);
# OK, now that we can check all region names. We will now assign a random value to each region:
rh_region_value_list = rnorm(length(atlas_region_names), 3.0, 1.0); # create 36 random values with mean 3 and stddev 1
names(rh_region_value_list) = atlas_region_names; # use the region names we retrieved earlier
# Now we have region_value_lists for both hemispheres. Time to visualize them:
vis.region.values.on.subject(template_subjects_dir, template_subject, atlas, lh_region_value_list, rh_region_value_list);
可视化时候,使用背景
subjects_dir = find.subjectsdir.of("fsaverage")$found_at;
subject_id = 'fsaverage';
lh_demo_cluster_file = system.file("extdata", "lh.clusters_fsaverage.mgz", package = "fsbrain", mustWork = TRUE);
rh_demo_cluster_file = system.file("extdata", "rh.clusters_fsaverage.mgz", package = "fsbrain", mustWork = TRUE);
lh_clust = freesurferformats::read.fs.morph(lh_demo_cluster_file); # contains a single positive cluster
rh_clust = freesurferformats::read.fs.morph(rh_demo_cluster_file); # contains two negative clusters
vis.symmetric.data.on.subject(subjects_dir, subject_id, lh_clust, rh_clust, bg="sulc");
输出
fsbrain.set.default.figsize(1200, 1200);
自己改编的代码,以适应dpabisurf的输出
subject_dir = 'Area_dataset/fsaverage5/' # folder generated by dpabisurf
subject_id = read.csv('subject_info.csv')$SubjectID
subject.l = list.files(subject_dir, pattern = "(hemi-L)", full.names = T) # read files
subject.r = list.files(subject_dir, pattern = "(hemi-R)", full.names = T)
# read function
Group_Morph_Standard = function(subject_file, subject_list){
# subject_file: L or R, should be defined early
# subject_list: subject ID, list
subject_data = list()
for(i in 1:length(subject_list)){
subject_data[[subject_list[i]]] = readgii(subject_file[i])[[1]][[1]][ ,1]
}
return(subject_data)
}
groupdata_stand = Group_Morph_Standard(subject.l, subject_list=subject_id)
补充-ubuntu安装fsbrain
安装fsbrain的时候,提示没有magick。
1.先去https://www.imagemagick.org/download/ImageMagick.tar.gz下载文件
2.解压 tar -xzvf ImageMagick-7.0.8-15; 进入目录 cd ImageMagick-7.0.8-15; 执行 ./configure,执行结束后在结尾可以看到已链接到tif
执行 make && make install
3.提示没有libdjvu/ddjvuapi.h no such file or directory。去下载libdjvu.
4.进入djvulibre-master文件夹,输入bash autogen.sh,然后sudo make install
5.重新进入ImageMagick,然后输入 sudo ./configure, sudo make && sudo make install
6.sudo ldconfig /usr/local/lib
这样,就安装好了ImageMagick了.
使用IDE的时候,报错“/lib/x86_64-linux-gnu/libstdc++.so.6”错误,这个是因为旧版本的libstdc占据了环境,新的libstdc没有导入。操作如下:
1.首先寻找,“sudo find / -name “libstdc++.so.6*””
2.可以看到最新的有miniconda的libstdc++.so.30
3.把这个libstdc++.so.30用sudo 复制到/lib/x86_64-linux-gnu/下面
4.删除原来的: sudo rm -rf libstdc++.so.6
5.制作软连接:sudo ln -s libstdc++.so.6.0.30 libstdc++.so.6
OK, enjoy it.