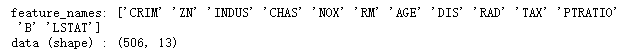3.1 机器学习 - 机器学习项目案例
机器学习 - 机器学习项目案例
案例1:利用岭回归研究波士顿房价
读取数据
from sklearn.datasets import load_boston
boston = load_boston()
print('feature_names:', boston.feature_names)
print('data (shape) :', boston.data.shape)
线性回归模型
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(boston.data, boston.target) # Fit
pre = lin_reg.predict(boston.data) # Predict
lin_reg.score(boston.data, boston.target) #Score
![]()
岭回归模型
from sklearn.linear_model import Ridge
ridge_reg = Ridge(alpha=0.5) # alpha值越大 正则化项所占比重越大
ridge_reg.fit(boston.data, boston.target) # Fit
ridge_reg.score(boston.data, boston.target) # Score
![]()
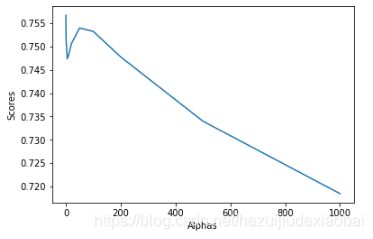
test_Ridge_alpha函数
探究alpha不同值,得到的回归结果
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
def test_Ridge_alpha(*data):
X_train, X_test, y_train, y_test = data
alphas = [0.01, 0.02, 0.05, 0.1, 0.2, 0.5, 1, 5, 10, 20, 50, 100, 200, 500, 1000]
scores = []
for i, alpha in enumerate(alphas):
ridge_reg = Ridge(alpha=alpha)
ridge_reg.fit(X_train, y_train)
scores.append(ridge_reg.score(X_test, y_test))
plt.xlabel('Alphas')
plt.ylabel('Scores')
sns.lineplot(x=alphas, y=scores)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(boston.data, boston.target, test_size=0.3, random_state=31)
test_Ridge_alpha(X_train, X_test, y_train, y_test)
invalid value encountered in true_divide # Remove the CWD from sys.path while we load stuff.
import numpy as np
np.seterr(divide='ignore', invalid='ignore')
案例2:利用决策树回归预测波士顿放假
import numpy as np
from sklearn.datasets import load_boston
from sklearn.model_selection import cross_val_score, cross_validate
from sklearn.tree import DecisionTreeRegressor
# Load DataSet
boston = load_boston()
X, y = boston.data, boston.target
features = boston.feature_names
# Fit
regression_tree = DecisionTreeRegressor(min_samples_split=30, min_samples_leaf=10, random_state=0) #决策树
regression_tree.fit(X, y)
# Score
score = np.mean(cross_val_score(regression_tree, X, y, cv=3)) # cv=3
print('Mean squared error: {0}'.format(round(abs(score),2)))
![]()
案例3:Logistic回归实现对鸢尾花数据分类
import matplotlib.pyplot as plt
from sklearn import datasets
iris = datasets.load_iris() # 加载鸢尾花数据
sepal_length_list = iris.data[:, 0] # 花萼长度
sepal_width_list = iris.data[:, 1] # 花萼宽度
# 构建 setosa、versicolor、virginica 索引数组
setosa_index_list = iris.target == 0 # setosa 索引数组
versicolor_index_list = iris.target == 1 # versicolor 索引数组
virginica_index_list = iris.target == 2 # virginica 索引数组
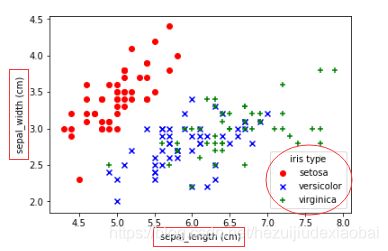
plt.scatter(sepal_length_list[setosa_index_list],
sepal_width_list[setosa_index_list], color="red", marker='o', label="setosa")
plt.scatter(sepal_length_list[versicolor_index_list],
sepal_width_list[versicolor_index_list], color="blue", marker="x", label="versicolor")
plt.scatter(sepal_length_list[virginica_index_list],
sepal_width_list[virginica_index_list],color="green", marker="+", label="virginica")
# 设置 legend
plt.legend(loc="best", title="iris type")
# 设定横坐标名称
plt.xlabel("sepal_length (cm)")
# 设定纵坐标名称
plt.ylabel("sepal_width (cm)")
逻辑回归
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn import datasets
# 加载鸢尾花数据
iris = datasets.load_iris()
# 设置训练集和测试集
X_train, X_test , y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.5, random_state=1)
# 创建一个Logistic回归分类器
logr = LogisticRegression(penalty='l2', random_state=0)
# 训练分类器
logr.fit(X_train, y_train)
# 预测所属类别
category = logr.predict(X_test)
category
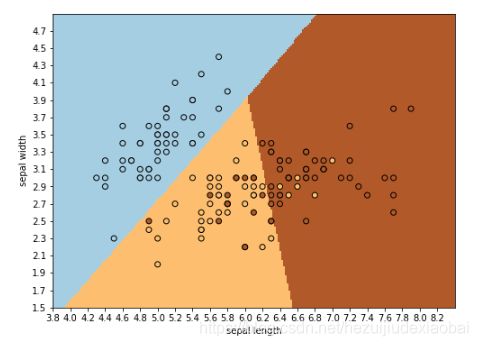
模型可视化
import numpy as np
import matplotlib.pyplot as plt
# 只考虑前两个特征,即花萼长度(sepal length)、花萼宽度(sepal width)
X = iris.data[:, 0:2]
y = iris.target
logreg = LogisticRegression(C=1e5) #C:惩罚项系数的倒数,越小,正则化项越大
logreg.fit(X, y)
# 网格大小
h = 0.02
x_min, x_max = X[:, 0].min() - 0.5, X[:, 0].max() + 0.5 # 将 X 的第一列(花萼长度)作为 x 轴,并求出 x 轴的最大值与最小值
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5 # 将 X 的第二列(花萼宽度)作为 y 轴,并求出 y 轴的最大值与最小值
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# 调用 ravel() 函数将 xx 和 yy 平铺,然后使用 np.c_ 将平铺后的列表拼接
# 生成需要预测的特征矩阵,每一行的表示一个样本,每一列表示每个特征的取值
pre_data = np.c_[xx.ravel(), yy.ravel()]
Z = logreg.predict(pre_data)
Z = Z.reshape(xx.shape)
plt.figure(1, figsize=(8, 6))
#
plt.pcolormesh(xx, yy, Z, cmap=plt.cm.Paired)
plt.scatter(X[:, 0], X[:, 1], c=y, edgecolors='k', cmap=plt.cm.Paired)
# 设置坐标轴label
plt.xlabel("sepal length")
plt.ylabel("sepal width")
# 设置坐标轴范围
plt.xlim(xx.min(), xx.max())
plt.ylim(yy.min(), yy.max())
# 设置坐标轴刻度
plt.xticks(np.arange(x_min, x_max, h * 10))
plt.yticks(np.arange(y_min, y_max, h * 10))
plt.show()
案例4:利用贝叶斯分类实现手写数字识别
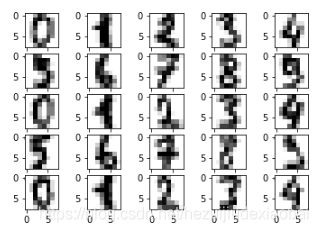
加载数据集
import matplotlib.pyplot as plt
from sklearn.datasets import load_digits
digits = load_digits()
fig = plt.figure()
for i in range(25):
ax = fig.add_subplot(5, 5, i+1)
ax.imshow(digits.images[i], cmap=plt.cm.gray_r, interpolation='nearest')
测试集的样本数
# 划分数据集
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(digits.data, digits.target, test_size=0.3, random_state=0)
# 测试集的样本数
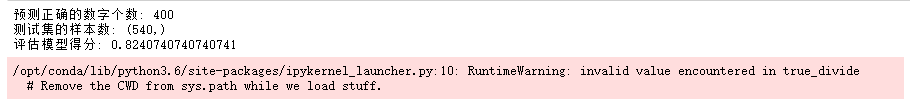
print("y_test(shape):", y_test.shape)
![]()
import numpy as np
np.seterr(divide='ignore', invalid='ignore')
![]()
GaussianNB
高斯贝叶斯分类器,特征的条件概率符合高斯分布
from sklearn.naive_bayes import GaussianNB
gau_nb = GaussianNB()
gau_nb.fit(X_train, y_train)
gy_pre = gau_nb.predict(X_test)
# 评估模型得分
print("Score:", gau_nb.score(X_test, y_test))
# 检验预测正确的数字个数
print("Right:", y_pre[(y_test / gy_pre) == 1].size)
MultinomialNB
多项式贝叶斯分类器,特征的条件概率符合多项式分布
from sklearn.naive_bayes import MultinomialNB
mul_nb = MultinomialNB()
mul_nb.fit(X_train, y_train)
my_pre = mul_nb.predict(X_test)
print("Score:", mul_nb.score(X_test, y_test))
print('Right:', my_pre[(y_test / my_pre) == 1].size)
BernoulliNB
伯努利贝叶斯分类器,符合伯努利分布(二项式分布)
from sklearn.naive_bayes import BernoulliNB
ber_nb = BernoulliNB()
ber_nb.fit(X_train, y_train)
by_pre = ber_nb.predict(X_test)
print("Score:", ber_nb.score(X_test, y_test))
print('Right:', by_pre[(y_test / by_pre) == 1].size)
模型可视化#1
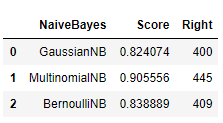
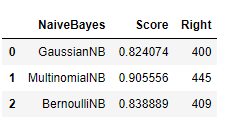
import pandas as pd
naive_bayes = pd.DataFrame(['GaussianNB', 'MultinomialNB', 'BernoulliNB'])
score = pd.DataFrame([gau_nb.score(X_test, y_test), mul_nb.score(X_test, y_test), ber_nb.score(X_test, y_test)])
right = pd.DataFrame([y_pre[(y_test / gy_pre) == 1].size, my_pre[(y_test / my_pre) == 1].size,
by_pre[(y_test / by_pre) == 1].size])
vs = pd.concat([naive_bayes, score, right], axis=1)
vs.columns = ['NaiveBayes', 'Score', 'Right']
vs
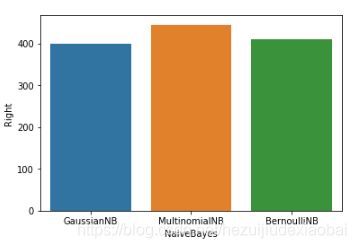
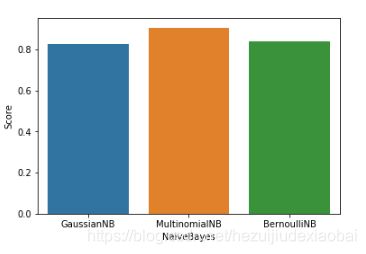
vs.plot.barh()
模型可视化#2
vs_naive_bayes = pd.DataFrame({'NaiveBayes': pd.Series(['GaussianNB', 'MultinomialNB', 'BernoulliNB']),
'Score': pd.Series([gau_nb.score(X_test, y_test), mul_nb.score(X_test, y_test), ber_nb.score(X_test, y_test)]),
'Right': pd.Series([y_pre[(y_test / gy_pre) == 1].size, my_pre[(y_test / my_pre) == 1].size, by_pre[(y_test / by_pre) == 1].size])})
vs_naive_bayes
import seaborn as sns
sns.barplot(vs_naive_bayes.NaiveBayes, vs_naive_bayes.Right)
sns.barplot(vs_naive_bayes.NaiveBayes, vs_naive_bayes.Score)
案例5:利用随机森林分类筛查乳腺癌
from sklearn.datasets import load_breast_cancer
dataset = load_breast_cancer()
dataset.target_names
![]()
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
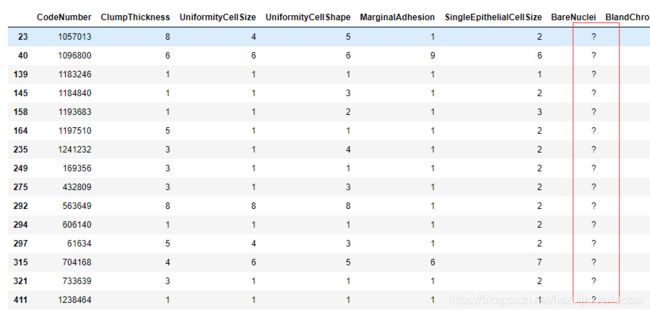
NAMES = ["CodeNumber", "ClumpThickness", "UniformityCellSize", "UniformityCellShape", "MarginalAdhesion", "SingleEpithelialCellSize", "BareNuclei", "BlandChromatin", "NormalNucleoli", "Mitoses", "CancerType"]
breast_cancer_data =pd.read_csv('http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data',
header=None,
names=NAMES)
breast_cancer_data
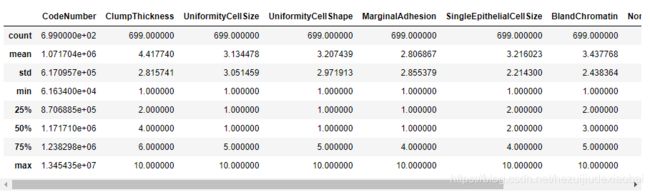
breast_cancer_data.describe()
train_x, test_x, train_y, test_y = train_test_split(breast_cancer_data[NAMES[1:-1]], breast_cancer_data[NAMES[-1]], train_size=0.7)
print("Train_x Shape :: ", train_x.shape)
print("Train_y Shape :: ", train_y.shape)
print("Test_x Shape :: ", test_x.shape)
print("Test_y Shape :: ", test_y.shape)
RandomForestClassifier #1
# 利用随机森林分类进行筛选
clf = RandomForestClassifier()
clf.fit(train_x, train_y)
predictions = clf.predict(test_x)
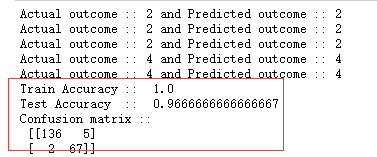
for i in range(0, 5):
print("Actual outcome :: {} and Predicted outcome :: {}".format(list(test_y)[i], predictions[i]))
print("Train Accuracy :: ", accuracy_score(train_y, clf.predict(train_x)))
print("Test Accuracy :: ", accuracy_score(test_y, predictions))
print(" Confusion matrix ", confusion_matrix(test_y, predictions))
breast_cancer_data.iloc[np.where(breast_cancer_data['BareNuclei'] == '?')]
# 计算异常值列的平均值
mean_value = breast_cancer_data[breast_cancer_data["BareNuclei"] != "?"]["BareNuclei"].astype(np.int).mean()
mean_value
breast_cancer_data['BareNuclei'] = breast_cancer_data['BareNuclei'].replace('?', mean_value) # mean_value替换?
breast_cancer_data.iloc[np.where(breast_cancer_data['BareNuclei'] == '?')]
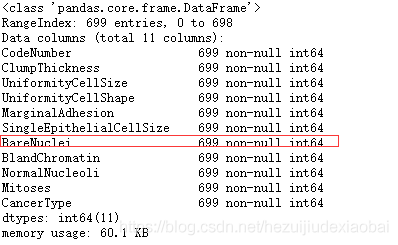
breast_cancer_data["BareNuclei"] = breast_cancer_data["BareNuclei"].astype(np.int64)
breast_cancer_data.info()
RandomForestClassifier #2
train_x, test_x, train_y, test_y = train_test_split(breast_cancer_data[NAMES[1:-1]], breast_cancer_data[NAMES[-1]], train_size=0.7)
# 利用随机森林分类进行筛选
clf = RandomForestClassifier()
clf.fit(train_x, train_y)
predictions = clf.predict(test_x)
for i in range(0, 5):
print("Actual outcome :: {} and Predicted outcome :: {}".format(list(test_y)[i], predictions[i]))
print("Train Accuracy :: ", accuracy_score(train_y, clf.predict(train_x)))
print("Test Accuracy :: ", accuracy_score(test_y, predictions))
print("Confusion matrix :: \n", confusion_matrix(test_y, predictions))
参考资料
DataFrame
Matplotlib
help(plt.pcolormesh)
Create a pseudocolor plot with a non-regular rectangular grid.
Numpy
help(np.meshgrid)
Return coordinate matrices from coordinate vectors.
help(np.ravel)
Return a contiguous flattened array.
help(np.c_)
Translates slice objects to concatenation along the second axis.
help(np.seterr)
Set how floating-point errors are handled.