图网络:从数据处理到DGL模型构建(GCN, GraphSAGE, RGCN)
目录
- 1.数据处理
-
- 1.1.原始数据节点去重
- 1.2.训练集和测试集的节点划分
- 1.3.边数据中删除节点
- 1.4.节点特征数据和Label
- 2.建图
-
- 2.1.同质图 Networkx→DGL
- 2.2.根据ID建图( 针对RGCN)
-
- 2.2.1.获得node和edge的ID
- 2.2.2.将原始边数据转换为ID数据
- 2.2.3.建图
- 3.节点特征, Label, 训练集/测试集idx
- 4.GCN→RF
-
- 4.1.模型结构
- 4.2.GCN函数定义
- 4.3.Step1:训练集训练GCN1
-
- 4.3.1.运行前获取数据
- 4.3.2.训练模型
- 4.4.Step2:获得测试集的graph embedding
-
- 4.4.1.运行前获取数据
- 4.4.2.运行模型
- 4.5.random forest分类
-
- 4.5.1.定义函数
- 4.5.2.数据准备
- 4.5.3.训练模型并预测
- 5.GraphSAGE
-
- 5.1.函数定义
- 5.2.参数设置
- 5.3.定义模型
- 5.4.训练
- 6.R-GCN
-
- 6.1.生成Edge norm用以建图
- 6.2.建图
- 6.3.函数定义
- 6.4.参数设置
- 6.5.训练和预测
- 6.6.加载模型并测试
- 7.学习资料
1.数据处理
1.1.原始数据节点去重
图网络学习和普通机器学习/深度学习算法在数据处理上是存在差异的。
任务:根据用户的行为(Feature),预测用户在下一个时间段的某行为(Label)。
原始数据:不同用户在不同时间节点下的Feature及相应的Label。
100个样本,其中仅有20个不同的用户。
在传统算法中,上述数据可直接以100个样本及其对应Label进行模型的训练。
但是在图算法中,仅可保留20个user作为20个样本。因为在图网络中,用户和节点是一一对应的。
本次实验直接采用去旧留新法,即只保留某user的最新日期的样本。
def obtain_node_data(file = 'train.txt'):
node_data = pd.read_table(file).sample(frac = 1.0)
node_data['vroleid'] = node_data['vroleid'].astype('str') # user id
print('node_data: shape = {}, # user = {}'.format(node_data.shape, len(set(node_data['vroleid']))))
node_data = node_data.dropna(axis = 1, how = 'all')
print('node_data: shape = {}, # user = {}'.format(node_data.shape, len(set(node_data['vroleid']))))
node_data = node_data.sort_values('stat_date', ascending = False).groupby('vroleid', as_index = False).first()
print('node_data: shape = {}, # user = {}'.format(node_data.shape, len(set(node_data['vroleid']))))
return node_data
1.2.训练集和测试集的节点划分
在实际应用中,会出现训练集和测试集的节点交叉的情形。
某user在训练集中是以20200101-20200201的时间段出现的。
该user在测试集是以20200301-20200401时间段出现的。
在本次实验中,对于此类交叉节点的处理是:只保留测试集中的最新时间的样本,用于测试。在训练集中去掉相关user的所有数据,以保证数据不重叠。
train_node_data = obtain_node_data('train.txt')
test_node_data = obtain_node_data('test.txt')
node_data = pd.concat([train_node_data, test_node_data])
node_data = node_data.sort_values('stat_date', ascending = False).groupby('vroleid', as_index = False).first()
print('node_data: shape = {}, # user = {}'.format(node_data.shape, len(set(node_data['vroleid']))))
train_node_data = node_data[~node_data['vroleid'].isin(list(test_node_data['vroleid']))]
test_node_data = node_data[node_data['vroleid'].isin(list(test_node_data['vroleid']))]
print('Shape: Train = {} | Test = {}'.format(train_node_data.shape, test_node_data.shape))
# 将训练集和测试集的节点数据合并(注意以train_mask和test_mask记录哪条节点数据属于哪个数据集。
train_mask_num = train_node_data.shape[0]
test_mask_num = test_node_data.shape[0]
print('Split Num: # Train = {}, # Test = {}'.format(train_mask_num, test_mask_num))
split_idx = [0] * train_mask_num + [1] * test_mask_num
node_data = pd.concat([train_node_data, test_node_data]).reset_index(drop = True)
node_data['split_idx'] = split_idx
1.3.边数据中删除节点
- 去掉未在节点数据中出现的节点(因为没有相对应的Label)。
def process_edge_data(edge_data, node_data = test_node_data):
edge_data = edge_data[edge_data['vroleid'].isin(node_data['vroleid'])]
edge_data = edge_data[edge_data['friend_roleid'].isin(node_data['vroleid'])]
return edge_data
edge_data = process_edge_data(edge_data, node_data = node_data)
- 根据图算法的需要:
- Transductive learning(如GCN)
- Inductive learning(如GraphSAGE)
Inductive learning:只根据现有的ABC来训练模型,在来一个新的数据时,直接加载5个ABC训练好的模型来预测。
Transductive learning:直接以某种算法观察出数据的分布,这里呈现三个cluster,就根据cluster判定,不会建立一个预测的模型。如果一个新的数据加进来,就必须重新算一遍整个算法,新加的数据也会导致旧的已预测?的结果改变。
参考:https://www.zhihu.com/question/68275921/answer/480709225
1.4.节点特征数据和Label
建图后才能处理。转Section 3。
2.建图
2.1.同质图 Networkx→DGL
-
先得到networkx的Graph
networkx.convert_matrix.from_pandas_edgelist
G = nx.from_pandas_edgelist(edge_data, 'vroleid', 'friend_roleid') print(nx.info(G)) # 将node_data中没有好友关系/边的user节点加入到G中,即不存在于edge_data中的node no_edge_node = set(train_node_data.index).difference(set(G.nodes)) G.add_nodes_from(no_edge_node) print(nx.info(G))注意: 原始的Edge数据可能并不能覆盖Node数据中的所有节点,因此对于未被覆盖的结点,要注意在建图的时候加进去,作为一个度为0的节点。
-
将networkx.graph转为DGL的Graph
g = dgl.DGLGraph() g.from_networkx(G) g.add_edges(g.nodes(), g.nodes()) # self-loop # g边数 = G中的边数*2 + len(g.nodes()) # 不建议使用这种方法,dgl.DGLGraph()将在后续版本中被淘汰。 # https://docs.dgl.ai/en/latest/api/python/graph.html#adding-nodes-and-edges# https://docs.dgl.ai/en/latest/generated/dgl.graph.html # 建议使用这种方法建图,因为上个版本将被淘汰 # 视算法情况决定是否加self-loop g = dgl.graph(G) print(g) # g边数 = G边数 * 2,可能是因为dgl.graph采用的是directed graph。
2.2.根据ID建图( 针对RGCN)
2.2.1.获得node和edge的ID
all_nodes = list(node_data['vroleid'])
entity2id = dict(zip(all_nodes, range(0, len(all_nodes))))
relation2id = dict(zip(range(1, 150), range(0, 149))) # 一共149个边类型(friend_level)
2.2.2.将原始边数据转换为ID数据
原始边数据:(vroleid, friend_level, friend_roleid)
转换为:(entity_id, relation_id, entity_id)
def read_triplets(edge_data, entity2id, relation2id):
edge_data = np.array(edge_data[['vroleid', 'friend_level', 'friend_roleid']])
triplets = []
for line in edge_data:
triplets.append((entity2id[line[0]], relation2id[line[1]], entity2id[line[-1]]))
return np.array(triplets)
all_triplets = read_triplets(edge_data[['vroleid', 'friend_roleid', 'friend_level']], entity2id, relation2id)
2.2.3.建图
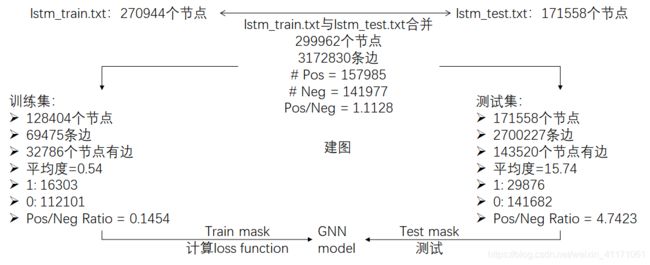
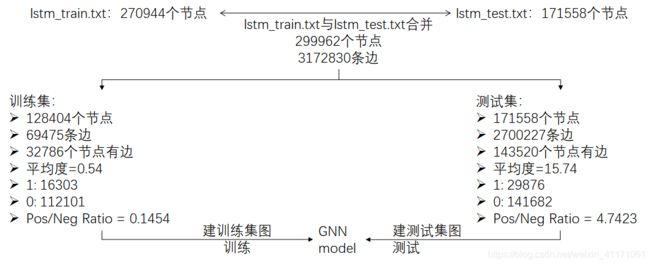
g = dgl.graph((all_triplets[:,0], all_triplets[:,2]), num_nodes = 299962)
3.节点特征, Label, 训练集/测试集idx
# 从node_data里提取和g.nodes()的顺序一致的Feature https://www.jianshu.com/p/2d3dd3e30d51
user_order = list(G.nodes()) # 针对Section 2.1
user_order = all_nodes # 针对Section 2.2
feature = deepcopy(node_data).reset_index()
feature['vroleid'] = feature['vroleid'].astype('category')
feature['vroleid'].cat.reorder_categories(user_order, inplace = True)
feature.sort_values('vroleid', inplace = True)
feature.set_index('vroleid', inplace = True)
feature.drop(['stat_date', 'vopenid', 'not_lost', 'split_idx', 'label_log', 'label_reg'], axis = 1, inplace = True)
# 标准化 https://scikit-learn.org/stable/modules/preprocessing.html#preprocessing
scaler = preprocessing.StandardScaler().fit(feature)
feature = scaler.transform(feature)
for i,j in zip(*np.where(np.isnan(feature))):
feature[i, j] = 0
labels = np.array(feature['not_lost'].astype(np.int))
label_log = np.array(feature['label_log'])
label_reg = np.array(feature['label_reg'])
split_mask_idx = np.array(feature['split_idx'].astype(np.int))
train_idx = np.where(split_mask_idx == 0)[0]
test_idx = np.where(split_mask_idx == 1)[0]
4.GCN→RF
4.1.模型结构
4.2.GCN函数定义
class GCNLayer(nn.Module):
def __init__(self, in_feats, out_feats):
super(GCNLayer, self).__init__()
self.linear = nn.Linear(in_feats, out_feats)
def forward(self, g, feature):
# Creating a local scope so that all the stored ndata and edata
# (such as the `'h'` ndata below) are automatically popped out
# when the scope exits.
with g.local_scope():
g.ndata['h'] = feature
g.update_all(gcn_msg, gcn_reduce)
h = g.ndata['h']
return self.linear(h)
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.layer1 = GCNLayer(180, 64)
self.layer2 = GCNLayer(64, 180)
def forward(self, g, features):
output = F.relu(self.layer1(g, features))
x = self.layer2(g, output)
return x, output
def evaluate(model, g, features, labels, mask):
model.eval() # 测试模式
with th.no_grad(): # 关闭求导
logits = model(g, features) # 所有数据作前向传播
logits = logits[mask] # 取出相应数据集对应的部分
labels = labels[mask]
_, indices = th.max(logits, dim = 1) # 按行取argmax,得到预测的标签
correct = th.sum(indices == labels)
tn, fp, fn, tp = confusion_matrix(labels, indices).ravel() # y_true, y_pred
assert correct.item() * 1.0 / len(labels) == (tn+tp)/len(labels)
accuracy = (tn+tp)/len(labels)
pos_acc = tp/sum(labels).item()
neg_acc = tn/(len(indices)-sum(indices).item()) # [y_true=0 & y_pred=0] / y_pred=0
neg_recall = tn / (tn+fp) # [y_true=0 & y_pred=0] / y_true=0
roc_auc = roc_auc_score(labels, logits[:,1])
prec, reca, _ = precision_recall_curve(labels, logits[:,1])
aupr = auc(reca, prec)
return neg_recall, neg_acc, pos_acc, accuracy, roc_auc, aupr
4.3.Step1:训练集训练GCN1
4.3.1.运行前获取数据
# step1中的所有图信息都是由train_node_data生成的。
g_step1 = dgl.DGLGraph()
g_step1.from_networkx(G_step1)
g_step1.add_edges(g_step1.nodes(), g_step1.nodes())
feature_step1 = th.FloatTensor(feature_step1)
labels_step1 = th.LongTensor(labels_step1)
train_idx_step1 = np.where(split_mask_idx_step1 == 0)[0]
val_idx_step1 = np.where(split_mask_idx_step1 == 1)[0]
train_mask_step1 = deepcopy(split_mask_idx_step1)
train_mask_step1[train_idx_step1] = 1
train_mask_step1[val_idx_step1] = 0
train_mask_step1 = th.BoolTensor(train_mask_step1)
val_mask_step1 = deepcopy(split_mask_idx_step1)
val_mask_step1[train_idx_step1] = 0
val_mask_step1[val_idx_step1] = 1
val_mask_step1 = th.BoolTensor(val_mask_step1)
4.3.2.训练模型
net = Net()
gcn_msg = fn.copy_src(src = 'h', out = 'm')
gcn_reduce = fn.sum(msg = 'm', out = 'h')
optimizer = th.optim.Adam(net.parameters(), lr=1e-3)
dur = []
for epoch in range(1, 51): # 完整遍历一遍训练集, 一个epoch做一次更新
print(epoch, end = ',')
t0 = time.time()
net.train()
logits, output_step1 = net(g_step1, feature_step1) # 所有数据前向传播
logp = F.log_softmax(logits, 1)
loss = F.nll_loss(logp[train_mask_step1], labels_step1[train_mask_step1]) # 只选择训练节点进行监督,计算loss
optimizer.zero_grad() # 清空梯度
loss.backward() # 反向传播计算参数的梯度
optimizer.step() # 使用优化方法进行梯度更新
dur.append(time.time() - t0)
4.4.Step2:获得测试集的graph embedding
4.4.1.运行前获取数据
# step2的所有数据都是由node_data得到的。
g_step2 = dgl.DGLGraph()
g_step2.from_networkx(G_step2)
g_step2.add_edges(g_step2.nodes(), g_step2.nodes())
feature_step2 = th.FloatTensor(feature_step2)
labels_step2 = th.LongTensor(labels_step2)
train_idx_step2 = np.where(split_mask_idx_step2 == 0)[0]
test_idx_step2 = np.where(split_mask_idx_step2 == 1)[0]
train_mask_step2 = deepcopy(split_mask_idx_step2)
train_mask_step2[train_idx_step2] = 1
train_mask_step2[test_idx_step2] = 0
train_mask_step2 = th.BoolTensor(train_mask_step2)
test_mask_step2 = deepcopy(split_mask_idx_step2)
test_mask_step2[train_idx_step2] = 0
test_mask_step2[test_idx_step2] = 1
test_mask_step2 = th.BoolTensor(test_mask_step2)
4.4.2.运行模型
optimizer = th.optim.Adam(net.parameters(), lr=1e-3)
dur = []
for epoch in range(1, 51): # 完整遍历一遍训练集, 一个epoch做一次更新
print(epoch, end = ',')
t0 = time.time()
net.train()
logits, output_step2 = net(g_step2, feature_step2) # 所有数据前向传播
logp = F.log_softmax(logits, 1)
loss = F.nll_loss(logp[train_mask_step2], labels_step2[train_mask_step2]) # 只选择训练节点进行监督,计算loss
optimizer.zero_grad() # 清空梯度
loss.backward() # 反向传播计算参数的梯度
optimizer.step() # 使用优化方法进行梯度更新
dur.append(time.time() - t0)
_, output_step2 = net(g_step2, feature_step2)
# 需要的是output_step2[test_mask_step2]
4.5.random forest分类
4.5.1.定义函数
def performance_evaluation(y_true, y_pred):
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel() # y_true, y_pred
accuracy = (tn+tp)/len(y_true)
pos_acc = tp/sum(y_true).item()
neg_acc = tn/(len(y_pred)-sum(y_pred).item()) # [y_true=0 & y_pred=0] / y_pred=0
neg_recall = tn / (tn+fp) # [y_true=0 & y_pred=0] / y_true=0
return neg_recall, neg_acc, pos_acc, accuracy
4.5.2.数据准备
x_step1 = output_step1.data.numpy()
y_step1 = labels_step1.data.numpy()
x_test = output_step2[test_mask_step2].data.numpy()
y_test = labels_step2[test_mask_step2].data.numpy()
4.5.3.训练模型并预测
clf = RandomForestClassifier(random_state = 0)
clf.fit(x_step1[train_idx_step1], y_step1[train_idx_step1])
y_train_pred = clf.predict(x_step1[train_idx_step1])
y_val_pred = clf.predict(x_step1[val_idx_step1])
y_test_pred = clf.predict(x_test)
y_train_proba = clf.predict_proba(x_step1[train_idx_step1])
y_val_proba = clf.predict_proba(x_step1[val_idx_step1])
y_test_proba = clf.predict_proba(x_test)
train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy = performance_evaluation(y_step1[train_idx_step1], y_train_pred)
val_neg_recall, val_neg_acc, val_pos_acc, val_accuracy = performance_evaluation(y_step1[val_idx_step1], y_val_pred)
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy = performance_evaluation(y_test, y_test_pred)
print('Train: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy))
print('Valid: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(val_neg_recall, val_neg_acc, val_pos_acc, val_accuracy))
print('Test: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy))
5.GraphSAGE
DGL实现
5.1.函数定义
class GraphSAGE(nn.Module):
def __init__(self,
in_feats,
n_hidden,
n_classes,
n_layers,
activation,
dropout):
super().__init__()
self.n_layers = n_layers
self.n_hidden = n_hidden
self.n_classes = n_classes
self.layers = nn.ModuleList()
self.layers.append(SAGEConv(in_feats, n_hidden, 'mean'))
for i in range(1, n_layers - 1):
self.layers.append(dglnn.SAGEConv(n_hidden, n_hidden, 'mean'))
self.layers.append(SAGEConv(n_hidden, n_classes, 'mean'))
self.dropout = nn.Dropout(dropout)
self.activation = activation
def forward(self, blocks, x):
# block 是我们采样获得的二部图,这里用于消息传播
# x 为节点特征
h = x
for l, (layer, block) in enumerate(zip(self.layers, blocks)):
h_dst = h[:block.number_of_dst_nodes()]
h = layer(block, (h, h_dst))
if l != len(self.layers) - 1:
h = self.activation(h)
h = self.dropout(h)
return h
def inference(self, g, x, batch_size, device):
# inference 用于评估测试,针对的是完全图
# 目前会出现重复计算的问题,优化方案还在 to do list 上
nodes = th.arange(g.number_of_nodes())
for l, layer in enumerate(self.layers):
y = th.zeros(g.number_of_nodes(),
self.n_hidden if l != len(self.layers) - 1 else self.n_classes)
for start in trange(0, len(nodes), batch_size):
end = start + batch_size
batch_nodes = nodes[start:end]
block = dgl.to_block(dgl.in_subgraph(g, batch_nodes), batch_nodes)
input_nodes = block.srcdata[dgl.NID]
h = th.Tensor(x[input_nodes]).to(device)
h_dst = h[:block.number_of_dst_nodes()]
h = layer(block, (h, h_dst))
if l != len(self.layers) - 1:
h = self.activation(h)
h = self.dropout(h)
y[start:end] = h.cpu()
x = y
return y
class NeighborSampler(object):
def __init__(self, g, fanouts):
"""
g 为 DGLGraph;
fanouts 为采样节点的数量,实验使用 10,25,指一阶邻居采样 10 个,二阶邻居采样 25 个。
"""
self.g = g
self.fanouts = fanouts
def sample_blocks(self, seeds):
seeds = th.LongTensor(np.asarray(seeds))
blocks = []
for fanout in self.fanouts:
# sample_neighbors 可以对每一个种子的节点进行邻居采样并返回相应的子图
# replace=True 表示用采样后的邻居节点代替所有邻居节点
frontier = dgl.sampling.sample_neighbors(g, seeds, fanout, replace=True)
# 将图转变为可以用于消息传递的二部图(源节点和目的节点)
# 其中源节点的 id 也可能包含目的节点的 id(原因上面说了)
# 转变为二部图主要是为了方便进行消息传递
block = dgl.to_block(frontier, seeds)
# 获取新图的源节点作为种子节点,为下一层作准备
# 之所以是从 src 中获取种子节点,是因为采样操作相对于聚合操作来说是一个逆向操作
seeds = block.srcdata[dgl.NID]
# 把这一层放在最前面。
# PS:如果数据量大的话,插入操作是不是不太友好。
blocks.insert(0, block)
return blocks
def compute_acc(pred, labels): # 计算准确率
y_pred = th.argmax(pred, dim=1) # 按行取argmax,得到预测的标签
tn, fp, fn, tp = confusion_matrix(labels, y_pred).ravel() # y_true, y_pred
accuracy = (tn+tp)/len(labels)
pos_acc = tp/sum(labels).item()
neg_acc = tn/(len(y_pred)-sum(y_pred).item()) # [y_true=0 & y_pred=0] / y_pred=0
neg_recall = tn / (tn+fp) # [y_true=0 & y_pred=0] / y_true=0
return neg_recall, neg_acc, pos_acc, accuracy
def evaluate(model, g, inputs, labels, val_mask, batch_size, device):
"""
评估模型,调用 model 的 inference 函数
"""
model.eval()
with th.no_grad():
pred = model.inference(g, inputs, batch_size, device)
model.train()
return compute_acc(pred[val_mask], labels[val_mask])
def load_subtensor(g, labels, seeds, input_nodes, device):
"""
将一组节点的特征和标签复制到 GPU 上。
"""
batch_inputs = th.Tensor(g.ndata['features'][input_nodes]).to(device)
batch_labels = labels[seeds].to(device)
return batch_inputs, batch_labels
5.2.参数设置
features = th.FloatTensor(feature)
labels = th.LongTensor(labels)
train_mask = deepcopy(split_mask_idx)
train_mask[train_idx] = 1
train_mask[test_idx] = 0
train_mask = th.BoolTensor(train_mask)
test_mask = deepcopy(split_mask_idx)
test_mask[train_idx] = 0
test_mask[test_idx] = 1
test_mask = th.BoolTensor(test_mask)
# 参数设置
in_feats = feature.shape[1] # 输入维度
n_classes = 2 # label的种类数
gpu = -1
num_epochs = 50
num_hidden = 64
num_layers = 2
fan_out = '5,5'
batch_size = 1024
log_every = 20 # 记录日志的频率
eval_every = 2
lr = 0.001
dropout = 0
num_workers = 0 # 用于采样进程的数量
if gpu >= 0:
device = th.device('cuda:%d' % gpu)
else:
device = th.device('cpu')
5.3.定义模型
gcn_msg = fn.copy_src(src = 'h', out = 'm')
gcn_reduce = fn.sum(msg = 'm', out = 'h')
# Create PyTorch DataLoader for constructing blocks
# collate_fn 参数指定了 sampler,可以对 batch 中的节点进行采样
sampler = NeighborSampler(g, [int(fanout) for fanout in fan_out.split(',')])
dataloader = DataLoader(
dataset = train_idx,
batch_size = batch_size,
collate_fn = sampler.sample_blocks,
shuffle = True,
drop_last = False,
num_workers = num_workers)
model = GraphSAGE(in_feats, num_hidden, n_classes, num_layers, F.relu, dropout)
model = model.to(device)
loss_fcn = nn.CrossEntropyLoss()
loss_fcn = loss_fcn.to(device)
optimizer = optim.Adam(model.parameters(), lr = lr)
5.4.训练
# Training loop
avg = 0
iter_tput = []
for epoch in range(num_epochs):
tic = time.time()
for step, blocks in enumerate(dataloader):
tic_step = time.time()
input_nodes = blocks[0].srcdata[dgl.NID]
seeds = blocks[-1].dstdata[dgl.NID]
# Load the input features as well as output labels
batch_inputs, batch_labels = load_subtensor(g, labels, seeds, input_nodes, device)
# Compute loss and prediction
batch_pred = model(blocks, batch_inputs)
loss = loss_fcn(batch_pred, batch_labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
iter_tput.append(len(seeds) / (time.time() - tic_step))
if step % log_every == 0:
train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy = compute_acc(batch_pred, batch_labels)
gpu_mem_alloc = th.cuda.max_memory_allocated() / 1000000 if th.cuda.is_available() else 0
print('Epoch {:05d} | Step {:05d} | Loss {:.4f} | Speed (samples/sec) {:.4f} | GPU {:.1f} MiB'.format(
epoch, step, loss.item(), np.mean(iter_tput[3:]), gpu_mem_alloc))
print('Train: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy))
toc = time.time()
print('Epoch Time(s): {:.4f}'.format(toc - tic))
if epoch >= 5:
avg += toc - tic
if epoch % eval_every == 0 and epoch != 0:
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy = evaluate(model, g, g.ndata['features'], labels, test_mask, batch_size, device)
print('Test: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy))
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy = evaluate(model, g, g.ndata['features'], labels, test_mask, batch_size, device)
print('Test: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f}'.format(test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy))
print('Avg epoch time: {}'.format(avg / epoch))
6.R-GCN
DGL实现:加node feature的minibatch版本
6.1.生成Edge norm用以建图
# edge_data是vroleid, friend_level, friend_roleid
# u,v=src,dst; eid是edge_data中的第几行
edge_norm = np.zeros(edge_data.shape[0])
for e in tqdm(range(1, 150)):
eid = np.array(edge_data[edge_data['friend_level'] == e].index)
u = th.Tensor(all_triplets[eid, 0])
v = th.Tensor(all_triplets[eid, 2])
_, inverse_index, count = th.unique(v, return_inverse=True, return_counts=True)
degrees = count[inverse_index]
norm = th.ones(eid.shape[0]) / degrees
norm = norm.unsqueeze(1)
edge_norm[eid] = np.array(norm).reshape(-1)
6.2.建图
g = dgl.graph((all_triplets[:,0], all_triplets[:,2]), num_nodes = 299962) # 前面建图的地方有写具体含义
g.ndata['_TYPE'] = th.Tensor([0]*299962).long()
g.edata['_TYPE'] = th.Tensor(all_triplets[:,1]).long()
g.ndata['_ID'] = th.Tensor(np.array(range(299962))).long()
g.edata['_ID'] = th.Tensor(np.array(range(len(all_triplets)))).long()
g.edata['norm'] = th.Tensor(edge_norm.reshape(edge_norm.shape[0],1))
category_id = 0 # len(g.ntypes) = 1
node_ids = th.arange(g.number_of_nodes())
node_tids = g.ndata[dgl.NTYPE]
loc = (node_tids == category_id)
target_idx = node_ids[loc]
target_idx.share_memory_()
node_feats = []
node_feats.append(th.Tensor(feature).share_memory_())
6.3.函数定义
import argparse
import itertools
import numpy as np
import time
import torch as th
import torch.nn as nn
import torch.nn.functional as F
import torch.multiprocessing as mp
from torch.multiprocessing import Queue
from torch.nn.parallel import DistributedDataParallel
from torch.utils.data import DataLoader
import dgl
from dgl import DGLGraph
from functools import partial
from dgl.nn import RelGraphConv
from _thread import start_new_thread
def compute_acc(pred, labels): # 计算准确率
y_pred = th.argmax(pred, dim=1).cpu() # 按行取argmax,得到预测的标签
labels = labels.cpu()
tn, fp, fn, tp = confusion_matrix(labels, y_pred).ravel() # y_true, y_pred
accuracy = (tn+tp)/len(labels)
pos_acc = tp/sum(labels).item()
neg_acc = tn/(len(y_pred)-sum(y_pred).item()) # [y_true=0 & y_pred=0] / y_pred=0
neg_recall = tn / (tn+fp) # [y_true=0 & y_pred=0] / y_true=0
return neg_recall, neg_acc, pos_acc, accuracy
class BaseRGCN(nn.Module):
def __init__(self, num_nodes, h_dim, out_dim, num_rels, num_bases,
num_hidden_layers=1, dropout=0,
use_self_loop=False, use_cuda=False):
super(BaseRGCN, self).__init__()
self.num_nodes = num_nodes
self.h_dim = h_dim
self.out_dim = out_dim
self.num_rels = num_rels
self.num_bases = None if num_bases < 0 else num_bases
self.num_hidden_layers = num_hidden_layers
self.dropout = dropout
self.use_self_loop = use_self_loop
self.use_cuda = use_cuda
# create rgcn layers
self.build_model()
def build_model(self):
self.layers = nn.ModuleList()
# i2h
i2h = self.build_input_layer()
if i2h is not None:
self.layers.append(i2h)
# h2h
for idx in range(self.num_hidden_layers):
h2h = self.build_hidden_layer(idx)
self.layers.append(h2h)
# h2o
h2o = self.build_output_layer()
if h2o is not None:
self.layers.append(h2o)
def build_input_layer(self):
return None
def build_hidden_layer(self, idx):
raise NotImplementedError
def build_output_layer(self):
return None
def forward(self, g, h, r, norm):
for layer in self.layers:
h = layer(g, h, r, norm)
return h
class RelGraphEmbedLayer(nn.Module):
r"""Embedding layer for featureless heterograph.
Parameters
----------
dev_id : int
Device to run the layer.
num_nodes : int
Number of nodes.
node_tides : tensor
Storing the node type id for each node starting from 0
num_of_ntype : int
Number of node types
input_size : list of int
A list of input feature size for each node type. If None, we then
treat certain input feature as an one-hot encoding feature.
embed_size : int
Output embed size
embed_name : str, optional
Embed name
"""
def __init__(self,
dev_id,
num_nodes,
node_tids,
num_of_ntype,
input_size,
embed_size,
sparse_emb=False,
embed_name='embed'):
super(RelGraphEmbedLayer, self).__init__()
self.dev_id = dev_id
self.embed_size = embed_size
self.embed_name = embed_name
self.num_nodes = num_nodes
self.sparse_emb = sparse_emb
# create weight embeddings for each node for each relation
self.embeds = nn.ParameterDict()
self.num_of_ntype = num_of_ntype
self.idmap = th.empty(num_nodes).long()
for ntype in range(num_of_ntype):
if input_size[ntype] is not None:
input_emb_size = input_size[ntype].shape[1]
embed = nn.Parameter(th.Tensor(input_emb_size, self.embed_size))
nn.init.xavier_uniform_(embed)
self.embeds[str(ntype)] = embed
self.node_embeds = th.nn.Embedding(node_tids.shape[0], self.embed_size, sparse=self.sparse_emb)
nn.init.uniform_(self.node_embeds.weight, -1.0, 1.0)
def forward(self, node_ids, node_tids, type_ids, features):
"""Forward computation
Parameters
----------
node_ids : tensor
node ids to generate embedding for.
node_ids : tensor
node type ids
features : list of features
list of initial features for nodes belong to different node type.
If None, the corresponding features is an one-hot encoding feature,
else use the features directly as input feature and matmul a
projection matrix.
Returns
-------
tensor
embeddings as the input of the next layer
"""
tsd_ids = node_ids.to(self.node_embeds.weight.device)
embeds = th.empty(node_ids.shape[0], self.embed_size, device=self.dev_id)
for ntype in range(self.num_of_ntype):
if features[ntype] is not None:
loc = node_tids == ntype
embeds[loc] = features[ntype][type_ids[loc]].to(self.dev_id) @ self.embeds[str(ntype)].to(self.dev_id)
else:
loc = node_tids == ntype
embeds[loc] = self.node_embeds(tsd_ids[loc]).to(self.dev_id)
return embeds
class EntityClassify(nn.Module):
""" Entity classification class for RGCN
Parameters
----------
device : int
Device to run the layer.
num_nodes : int
Number of nodes.
h_dim : int
Hidden dim size.
out_dim : int
Output dim size.
num_rels : int
Numer of relation types.
num_bases : int
Number of bases. If is none, use number of relations.
num_hidden_layers : int
Number of hidden RelGraphConv Layer
dropout : float
Dropout
use_self_loop : bool
Use self loop if True, default False.
low_mem : bool
True to use low memory implementation of relation message passing function
trade speed with memory consumption
"""
def __init__(self,
device,
num_nodes,
h_dim,
out_dim,
num_rels,
num_bases=None,
num_hidden_layers=1,
dropout=0,
use_self_loop=False,
low_mem=False,
layer_norm=False):
super(EntityClassify, self).__init__()
self.device = th.device(device if device >= 0 else 'cpu')
self.num_nodes = num_nodes
self.h_dim = h_dim
self.out_dim = out_dim
self.num_rels = num_rels
self.num_bases = None if num_bases < 0 else num_bases
self.num_hidden_layers = num_hidden_layers
self.dropout = dropout
self.use_self_loop = use_self_loop
self.low_mem = low_mem
self.layer_norm = layer_norm
self.layers = nn.ModuleList()
# i2h
self.layers.append(RelGraphConv(
self.h_dim, self.h_dim, self.num_rels, "basis",
self.num_bases, activation=F.relu, self_loop=self.use_self_loop,
low_mem=self.low_mem, dropout=self.dropout))
# h2h
for idx in range(self.num_hidden_layers):
self.layers.append(RelGraphConv(
self.h_dim, self.h_dim, self.num_rels, "basis",
self.num_bases, activation=F.relu, self_loop=self.use_self_loop,
low_mem=self.low_mem, dropout=self.dropout))
# h2o
self.layers.append(RelGraphConv(
self.h_dim, self.out_dim, self.num_rels, "basis",
self.num_bases, activation=None,
self_loop=self.use_self_loop,
low_mem=self.low_mem))
def forward(self, blocks, feats, norm=None):
if blocks is None:
# full graph training
blocks = [self.g] * len(self.layers)
h = feats
# print('1: ', h.shape)
for layer, block in zip(self.layers, blocks):
block = block.to(self.device)
# print('2: ', h.shape)
h = layer(block, h, block.edata['etype'], block.edata['norm'])
# print('3: ', h.shape)
return h
class NeighborSampler:
"""Neighbor sampler
Parameters
----------
g : DGLHeterograph
Full graph
target_idx : tensor
The target training node IDs in g
fanouts : list of int
Fanout of each hop starting from the seed nodes. If a fanout is None,
sample full neighbors.
"""
def __init__(self, g, target_idx, fanouts):
self.g = g
self.target_idx = target_idx
self.fanouts = fanouts
"""Do neighbor sample
Parameters
----------
seeds :
Seed nodes
Returns
-------
tensor
Seed nodes, also known as target nodes
blocks
Sampled subgraphs
"""
def sample_blocks(self, seeds):
blocks = []
etypes = []
norms = []
ntypes = []
seeds = th.tensor(seeds).long()
cur = self.target_idx[seeds]
for fanout in self.fanouts:
if fanout is None or fanout == -1:
frontier = dgl.in_subgraph(self.g, cur)
else:
frontier = dgl.sampling.sample_neighbors(self.g, cur, fanout)
etypes = self.g.edata[dgl.ETYPE][frontier.edata[dgl.EID]]
norm = self.g.edata['norm'][frontier.edata[dgl.EID]]
block = dgl.to_block(frontier, cur)
block.srcdata[dgl.NTYPE] = self.g.ndata[dgl.NTYPE][block.srcdata[dgl.NID]]
block.srcdata['type_id'] =self.g.ndata[dgl.NID][block.srcdata[dgl.NID]]
block.edata['etype'] = etypes
block.edata['norm'] = norm
cur = block.srcdata[dgl.NID]
blocks.insert(0, block)
return seeds, blocks
# https://github.com/classicsong/dgl/blob/a5d10b893877bf58dd9322804b8a552ffdbaf932/examples/pytorch/rgcn/utils.py
def get_adj_and_degrees(num_nodes, triplets):
""" Get adjacency list and degrees of the graph
"""
adj_list = [[] for _ in range(num_nodes)]
for i,triplet in enumerate(triplets):
adj_list[triplet[0]].append([i, triplet[2]])
adj_list[triplet[2]].append([i, triplet[0]])
degrees = np.array([len(a) for a in adj_list])
adj_list = [np.array(a) for a in adj_list]
return adj_list, degrees
def sample_edge_neighborhood(adj_list, degrees, n_triplets, sample_size):
"""Sample edges by neighborhool expansion.
This guarantees that the sampled edges form a connected graph, which
may help deeper GNNs that require information from more than one hop.
"""
edges = np.zeros((sample_size), dtype=np.int32)
#initialize
sample_counts = np.array([d for d in degrees])
picked = np.array([False for _ in range(n_triplets)])
seen = np.array([False for _ in degrees])
for i in range(0, sample_size):
weights = sample_counts * seen
if np.sum(weights) == 0:
weights = np.ones_like(weights)
weights[np.where(sample_counts == 0)] = 0
probabilities = (weights) / np.sum(weights)
chosen_vertex = np.random.choice(np.arange(degrees.shape[0]),
p=probabilities)
chosen_adj_list = adj_list[chosen_vertex]
seen[chosen_vertex] = True
chosen_edge = np.random.choice(np.arange(chosen_adj_list.shape[0]))
chosen_edge = chosen_adj_list[chosen_edge]
edge_number = chosen_edge[0]
while picked[edge_number]:
chosen_edge = np.random.choice(np.arange(chosen_adj_list.shape[0]))
chosen_edge = chosen_adj_list[chosen_edge]
edge_number = chosen_edge[0]
edges[i] = edge_number
other_vertex = chosen_edge[1]
picked[edge_number] = True
sample_counts[chosen_vertex] -= 1
sample_counts[other_vertex] -= 1
seen[other_vertex] = True
return edges
def sample_edge_uniform(adj_list, degrees, n_triplets, sample_size):
"""Sample edges uniformly from all the edges."""
all_edges = np.arange(n_triplets)
return np.random.choice(all_edges, sample_size, replace=False)
def generate_sampled_graph_and_labels(triplets, sample_size, split_size,
num_rels, adj_list, degrees,
negative_rate, sampler="uniform"):
"""Get training graph and signals
First perform edge neighborhood sampling on graph, then perform negative
sampling to generate negative samples
"""
# perform edge neighbor sampling
if sampler == "uniform":
edges = sample_edge_uniform(adj_list, degrees, len(triplets), sample_size)
elif sampler == "neighbor":
edges = sample_edge_neighborhood(adj_list, degrees, len(triplets), sample_size)
else:
raise ValueError("Sampler type must be either 'uniform' or 'neighbor'.")
# relabel nodes to have consecutive node ids
edges = triplets[edges]
src, rel, dst = edges.transpose()
uniq_v, edges = np.unique((src, dst), return_inverse=True)
src, dst = np.reshape(edges, (2, -1))
relabeled_edges = np.stack((src, rel, dst)).transpose()
# negative sampling
samples, labels = negative_sampling(relabeled_edges, len(uniq_v),
negative_rate)
# further split graph, only half of the edges will be used as graph
# structure, while the rest half is used as unseen positive samples
split_size = int(sample_size * split_size)
graph_split_ids = np.random.choice(np.arange(sample_size),
size=split_size, replace=False)
src = src[graph_split_ids]
dst = dst[graph_split_ids]
rel = rel[graph_split_ids]
# build DGL graph
print("# sampled nodes: {}".format(len(uniq_v)))
print("# sampled edges: {}".format(len(src) * 2))
g, rel, norm = build_graph_from_triplets(len(uniq_v), num_rels,
(src, rel, dst))
return g, uniq_v, rel, norm, samples, labels
def comp_deg_norm(g):
g = g.local_var()
in_deg = g.in_degrees(range(g.number_of_nodes())).float().numpy()
norm = 1.0 / in_deg
norm[np.isinf(norm)] = 0
return norm
def build_graph_from_triplets(num_nodes, num_rels, triplets):
""" Create a DGL graph. The graph is bidirectional because RGCN authors
use reversed relations.
This function also generates edge type and normalization factor
(reciprocal of node incoming degree)
"""
g = dgl.DGLGraph()
g.add_nodes(num_nodes)
src, rel, dst = triplets
src, dst = np.concatenate((src, dst)), np.concatenate((dst, src))
rel = np.concatenate((rel, rel + num_rels))
edges = sorted(zip(dst, src, rel))
dst, src, rel = np.array(edges).transpose()
g.add_edges(src, dst)
norm = comp_deg_norm(g)
print("# nodes: {}, # edges: {}".format(num_nodes, len(src)))
return g, rel.astype('int64'), norm.astype('int64')
def build_test_graph(num_nodes, num_rels, edges):
src, rel, dst = edges.transpose()
print("Test graph:")
return build_graph_from_triplets(num_nodes, num_rels, (src, rel, dst))
def negative_sampling(pos_samples, num_entity, negative_rate):
size_of_batch = len(pos_samples)
num_to_generate = size_of_batch * negative_rate
neg_samples = np.tile(pos_samples, (negative_rate, 1))
labels = np.zeros(size_of_batch * (negative_rate + 1), dtype=np.float32)
labels[: size_of_batch] = 1
values = np.random.randint(num_entity, size=num_to_generate)
choices = np.random.uniform(size=num_to_generate)
subj = choices > 0.5
obj = choices <= 0.5
neg_samples[subj, 0] = values[subj]
neg_samples[obj, 2] = values[obj]
return np.concatenate((pos_samples, neg_samples)), labels
#######################################################################
#
# Utility functions for evaluations (raw)
#
#######################################################################
def sort_and_rank(score, target):
_, indices = torch.sort(score, dim=1, descending=True)
indices = torch.nonzero(indices == target.view(-1, 1))
indices = indices[:, 1].view(-1)
return indices
def perturb_and_get_raw_rank(embedding, w, a, r, b, test_size, batch_size=100):
""" Perturb one element in the triplets
"""
n_batch = (test_size + batch_size - 1) // batch_size
ranks = []
for idx in range(n_batch):
print("batch {} / {}".format(idx, n_batch))
batch_start = idx * batch_size
batch_end = min(test_size, (idx + 1) * batch_size)
batch_a = a[batch_start: batch_end]
batch_r = r[batch_start: batch_end]
emb_ar = embedding[batch_a] * w[batch_r]
emb_ar = emb_ar.transpose(0, 1).unsqueeze(2) # size: D x E x 1
emb_c = embedding.transpose(0, 1).unsqueeze(1) # size: D x 1 x V
# out-prod and reduce sum
out_prod = torch.bmm(emb_ar, emb_c) # size D x E x V
score = torch.sum(out_prod, dim=0) # size E x V
score = torch.sigmoid(score)
target = b[batch_start: batch_end]
ranks.append(sort_and_rank(score, target))
return torch.cat(ranks)
# return MRR (raw), and Hits @ (1, 3, 10)
def calc_raw_mrr(embedding, w, test_triplets, hits=[], eval_bz=100):
with torch.no_grad():
s = test_triplets[:, 0]
r = test_triplets[:, 1]
o = test_triplets[:, 2]
test_size = test_triplets.shape[0]
# perturb subject
ranks_s = perturb_and_get_raw_rank(embedding, w, o, r, s, test_size, eval_bz)
# perturb object
ranks_o = perturb_and_get_raw_rank(embedding, w, s, r, o, test_size, eval_bz)
ranks = torch.cat([ranks_s, ranks_o])
ranks += 1 # change to 1-indexed
mrr = torch.mean(1.0 / ranks.float())
print("MRR (raw): {:.6f}".format(mrr.item()))
for hit in hits:
avg_count = torch.mean((ranks <= hit).float())
print("Hits (raw) @ {}: {:.6f}".format(hit, avg_count.item()))
return mrr.item()
#######################################################################
#
# Utility functions for evaluations (filtered)
#
#######################################################################
def filter_o(triplets_to_filter, target_s, target_r, target_o, num_entities):
target_s, target_r, target_o = int(target_s), int(target_r), int(target_o)
filtered_o = []
# Do not filter out the test triplet, since we want to predict on it
if (target_s, target_r, target_o) in triplets_to_filter:
triplets_to_filter.remove((target_s, target_r, target_o))
# Do not consider an object if it is part of a triplet to filter
for o in range(num_entities):
if (target_s, target_r, o) not in triplets_to_filter:
filtered_o.append(o)
return torch.LongTensor(filtered_o)
def filter_s(triplets_to_filter, target_s, target_r, target_o, num_entities):
target_s, target_r, target_o = int(target_s), int(target_r), int(target_o)
filtered_s = []
# Do not filter out the test triplet, since we want to predict on it
if (target_s, target_r, target_o) in triplets_to_filter:
triplets_to_filter.remove((target_s, target_r, target_o))
# Do not consider a subject if it is part of a triplet to filter
for s in range(num_entities):
if (s, target_r, target_o) not in triplets_to_filter:
filtered_s.append(s)
return torch.LongTensor(filtered_s)
def perturb_o_and_get_filtered_rank(embedding, w, s, r, o, test_size, triplets_to_filter):
""" Perturb object in the triplets
"""
num_entities = embedding.shape[0]
ranks = []
for idx in range(test_size):
if idx % 100 == 0:
print("test triplet {} / {}".format(idx, test_size))
target_s = s[idx]
target_r = r[idx]
target_o = o[idx]
filtered_o = filter_o(triplets_to_filter, target_s, target_r, target_o, num_entities)
target_o_idx = int((filtered_o == target_o).nonzero())
emb_s = embedding[target_s]
emb_r = w[target_r]
emb_o = embedding[filtered_o]
emb_triplet = emb_s * emb_r * emb_o
scores = torch.sigmoid(torch.sum(emb_triplet, dim=1))
_, indices = torch.sort(scores, descending=True)
rank = int((indices == target_o_idx).nonzero())
ranks.append(rank)
return torch.LongTensor(ranks)
def perturb_s_and_get_filtered_rank(embedding, w, s, r, o, test_size, triplets_to_filter):
""" Perturb subject in the triplets
"""
num_entities = embedding.shape[0]
ranks = []
for idx in range(test_size):
if idx % 100 == 0:
print("test triplet {} / {}".format(idx, test_size))
target_s = s[idx]
target_r = r[idx]
target_o = o[idx]
filtered_s = filter_s(triplets_to_filter, target_s, target_r, target_o, num_entities)
target_s_idx = int((filtered_s == target_s).nonzero())
emb_s = embedding[filtered_s]
emb_r = w[target_r]
emb_o = embedding[target_o]
emb_triplet = emb_s * emb_r * emb_o
scores = torch.sigmoid(torch.sum(emb_triplet, dim=1))
_, indices = torch.sort(scores, descending=True)
rank = int((indices == target_s_idx).nonzero())
ranks.append(rank)
return torch.LongTensor(ranks)
def calc_filtered_mrr(embedding, w, train_triplets, valid_triplets, test_triplets, hits=[]):
with torch.no_grad():
s = test_triplets[:, 0]
r = test_triplets[:, 1]
o = test_triplets[:, 2]
test_size = test_triplets.shape[0]
triplets_to_filter = torch.cat([train_triplets, valid_triplets, test_triplets]).tolist()
triplets_to_filter = {tuple(triplet) for triplet in triplets_to_filter}
print('Perturbing subject...')
ranks_s = perturb_s_and_get_filtered_rank(embedding, w, s, r, o, test_size, triplets_to_filter)
print('Perturbing object...')
ranks_o = perturb_o_and_get_filtered_rank(embedding, w, s, r, o, test_size, triplets_to_filter)
ranks = torch.cat([ranks_s, ranks_o])
ranks += 1 # change to 1-indexed
mrr = torch.mean(1.0 / ranks.float())
print("MRR (filtered): {:.6f}".format(mrr.item()))
for hit in hits:
avg_count = torch.mean((ranks <= hit).float())
print("Hits (filtered) @ {}: {:.6f}".format(hit, avg_count.item()))
return mrr.item()
#######################################################################
#
# Main evaluation function
#
#######################################################################
def calc_mrr(embedding, w, train_triplets, valid_triplets, test_triplets, hits=[], eval_bz=100, eval_p="filtered"):
if eval_p == "filtered":
mrr = calc_filtered_mrr(embedding, w, train_triplets, valid_triplets, test_triplets, hits)
else:
mrr = calc_raw_mrr(embedding, w, test_triplets, hits, eval_bz)
return mrr
#######################################################################
#
# Multithread wrapper
#
#######################################################################
# According to https://github.com/pytorch/pytorch/issues/17199, this decorator
# is necessary to make fork() and openmp work together.
def thread_wrapped_func(func):
"""
Wraps a process entry point to make it work with OpenMP.
"""
from functools import wraps
@wraps(func)
def decorated_function(*args, **kwargs):
queue = Queue()
def _queue_result():
exception, trace, res = None, None, None
try:
res = func(*args, **kwargs)
except Exception as e:
exception = e
trace = traceback.format_exc()
queue.put((res, exception, trace))
start_new_thread(_queue_result, ())
result, exception, trace = queue.get()
if exception is None:
return result
else:
assert isinstance(exception, Exception)
raise exception.__class__(trace)
return decorated_function
@thread_wrapped_func
def run(proc_id, n_gpus, args, devices, dataset, split, queue=None):
dev_id = devices[proc_id]
g, node_feats, num_of_ntype, num_classes, num_rels, target_idx, \
train_idx, val_idx, test_idx, labels = dataset
if split is not None:
train_seed, val_seed, test_seed = split
train_idx = train_idx[train_seed]
val_idx = val_idx[val_seed]
test_idx = test_idx[test_seed]
fanouts = [int(fanout) for fanout in args.fanout.split(',')]
node_tids = g.ndata[dgl.NTYPE]
sampler = NeighborSampler(g, target_idx, fanouts)
loader = DataLoader(dataset=train_idx.numpy(),
batch_size=args.batch_size,
collate_fn=sampler.sample_blocks,
shuffle=True,
num_workers=args.num_workers)
# validation sampler
val_sampler = NeighborSampler(g, target_idx, [None] * args.n_layers)
val_loader = DataLoader(dataset=val_idx.numpy(),
batch_size=args.eval_batch_size,
collate_fn=val_sampler.sample_blocks,
shuffle=False,
num_workers=args.num_workers)
# validation sampler
test_sampler = NeighborSampler(g, target_idx, [None] * args.n_layers)
test_loader = DataLoader(dataset=test_idx.numpy(),
batch_size=args.eval_batch_size,
collate_fn=test_sampler.sample_blocks,
shuffle=False,
num_workers=args.num_workers)
if n_gpus > 1:
dist_init_method = 'tcp://{master_ip}:{master_port}'.format(
master_ip='127.0.0.1', master_port='12345')
world_size = n_gpus
backend = 'nccl'
# using sparse embedding or usig mix_cpu_gpu model (embedding model can not be stored in GPU)
if args.sparse_embedding or args.mix_cpu_gpu:
backend = 'gloo'
th.distributed.init_process_group(backend=backend,
init_method=dist_init_method,
world_size=world_size,
rank=dev_id)
# node features
# None for one-hot feature, if not none, it should be the feature tensor.
#
embed_layer = RelGraphEmbedLayer(dev_id,
g.number_of_nodes(),
node_tids,
num_of_ntype,
node_feats,
args.n_hidden,
sparse_emb=args.sparse_embedding)
# create model
# all model params are in device.
model = EntityClassify(dev_id,
g.number_of_nodes(),
args.n_hidden,
num_classes,
num_rels,
num_bases=args.n_bases,
num_hidden_layers=args.n_layers - 2,
dropout=args.dropout,
use_self_loop=args.use_self_loop,
low_mem=args.low_mem,
layer_norm=args.layer_norm)
if dev_id >= 0 and n_gpus == 1:
th.cuda.set_device(dev_id)
labels = labels.to(dev_id)
model.cuda(dev_id)
# embedding layer may not fit into GPU, then use mix_cpu_gpu
if args.mix_cpu_gpu is False:
embed_layer.cuda(dev_id)
if n_gpus > 1:
labels = labels.to(dev_id)
model.cuda(dev_id)
if args.mix_cpu_gpu:
embed_layer = DistributedDataParallel(embed_layer, device_ids=None, output_device=None)
else:
embed_layer.cuda(dev_id)
embed_layer = DistributedDataParallel(embed_layer, device_ids=[dev_id], output_device=dev_id)
model = DistributedDataParallel(model, device_ids=[dev_id], output_device=dev_id)
# optimizer
if args.sparse_embedding:
dense_params = list(model.parameters())
if args.node_feats:
if n_gpus > 1:
dense_params += list(embed_layer.module.embeds.parameters())
else:
dense_params += list(embed_layer.embeds.parameters())
optimizer = th.optim.Adam(dense_params, lr=args.lr, weight_decay=args.l2norm)
if n_gpus > 1:
emb_optimizer = th.optim.SparseAdam(embed_layer.module.node_embeds.parameters(), lr=args.lr)
else:
emb_optimizer = th.optim.SparseAdam(embed_layer.node_embeds.parameters(), lr=args.lr)
else:
all_params = list(model.parameters()) + list(embed_layer.parameters())
optimizer = th.optim.Adam(all_params, lr=args.lr, weight_decay=args.l2norm)
# training loop
print("start training...")
forward_time = []
backward_time = []
for epoch in range(args.n_epochs):
model.train()
for i, sample_data in enumerate(loader):
seeds, blocks = sample_data
t0 = time.time()
if args.mix_cpu_gpu is False:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
else:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
logits = model(blocks, feats)
loss = F.cross_entropy(logits, labels[seeds])
t1 = time.time()
optimizer.zero_grad()
if args.sparse_embedding:
emb_optimizer.zero_grad()
loss.backward()
optimizer.step()
if args.sparse_embedding:
emb_optimizer.step()
t2 = time.time()
forward_time.append(t1 - t0)
backward_time.append(t2 - t1)
train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy = compute_acc(logits, labels[seeds])
print('Epoch {} |Sample = {}/{} |Neg Recall = {:.2f} |Neg Acc = {:.2f} |Pos Acc = {:.2f} |All Acc = {:.2f} |Loss = {:.2f}'.format(
epoch, i+1, len(loader), train_neg_recall, train_neg_acc, train_pos_acc, train_accuracy, loss.item()))
print("Epoch {:05d}:{:05d} | Train Forward Time(s) {:.4f} | Backward Time(s) {:.4f}".
format(epoch, i+1, forward_time[-1], backward_time[-1]))
th.save(model.state_dict(), './checkpoints/model_5.3_RGCN_nodefeature_mp.pt')
th.save(model, './checkpoints/model_5.3_RGCN_nodefeature_mp.pkl')
# # only process 0 will do the evaluation
# if (queue is not None) or (proc_id == 0):
# model.eval()
# eval_logits = []
# eval_seeds = []
# with th.no_grad():
# for sample_data in tqdm(val_loader):
# th.cuda.empty_cache()
# seeds, blocks = sample_data
# if args.mix_cpu_gpu is False:
# feats = embed_layer(blocks[0].srcdata[dgl.NID],
# blocks[0].srcdata[dgl.NTYPE],
# blocks[0].srcdata['type_id'],
# node_feats)
# else:
# feats = embed_layer(blocks[0].srcdata[dgl.NID],
# blocks[0].srcdata[dgl.NTYPE],
# blocks[0].srcdata['type_id'],
# node_feats)
# logits = model(blocks, feats)
# eval_logits.append(logits.cpu().detach())
# eval_seeds.append(seeds.cpu().detach())
# eval_logits = th.cat(eval_logits)
# eval_seeds = th.cat(eval_seeds)
# if queue is not None:
# queue.put((eval_logits, eval_seeds))
# if proc_id == 0:
# if queue is not None:
# eval_logits = []
# eval_seeds = []
# for i in range(n_gpus):
# log = queue.get()
# val_l, val_s = log
# eval_logits.append(val_l)
# eval_seeds.append(val_s)
# eval_logits = th.cat(eval_logits)
# eval_seeds = th.cat(eval_seeds)
# val_loss = F.cross_entropy(eval_logits, labels[eval_seeds].cpu()).item()
# val_acc = th.sum(eval_logits.argmax(dim=1) == labels[eval_seeds].cpu()).item() / len(eval_seeds)
# print("Validation Accuracy: {:.4f} | Validation loss: {:.4f}".
# format(val_acc, val_loss))
if n_gpus > 1:
th.distributed.barrier()
print('===========Test start==========')
# only process 0 will do the evaluation
if (queue is not None) or (proc_id == 0):
model.eval()
test_logits = []
test_seeds = []
with th.no_grad():
for sample_data in tqdm(test_loader):
th.cuda.empty_cache()
seeds, blocks = sample_data
if args.mix_cpu_gpu is False:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
else:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
logits = model(blocks, feats)
test_logits.append(logits.cpu().detach())
test_seeds.append(seeds.cpu().detach())
test_logits = th.cat(test_logits)
test_seeds = th.cat(test_seeds)
if queue is not None:
queue.put((test_logits, test_seeds))
if proc_id == 0:
if queue is not None:
test_logits = []
test_seeds = []
for i in range(n_gpus):
log = queue.get()
test_l, test_s = log
test_logits.append(test_l)
test_seeds.append(test_s)
test_logits = th.cat(test_logits)
test_seeds = th.cat(test_seeds)
test_loss = F.cross_entropy(test_logits, labels[test_seeds].cpu()).item()
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy = compute_acc(test_logits, labels[test_seeds])
print('Test: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f} | Loss = {:.4f}'.format(
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy, test_loss))
print()
# sync for test
if n_gpus > 1:
th.distributed.barrier()
print("{}/{} Mean forward time: {:4f}".format(proc_id, n_gpus,
np.mean(forward_time[len(forward_time) // 4:])))
print("{}/{} Mean backward time: {:4f}".format(proc_id, n_gpus,
np.mean(backward_time[len(backward_time) // 4:])))
6.4.参数设置
parser = argparse.ArgumentParser(description='RGCN')
parser.add_argument("--dropout", type=float, default=0,
help="dropout probability")
parser.add_argument("--n-hidden", type=int, default=64,
help="number of hidden units")
parser.add_argument("--gpu", type=str, default='0',
help="gpu")
parser.add_argument("--lr", type=float, default=1e-2,
help="learning rate")
parser.add_argument("--n-bases", type=int, default=-1,
help="number of filter weight matrices, default: -1 [use all]")
parser.add_argument("--n-layers", type=int, default=2,
help="number of propagation rounds")
parser.add_argument("-e", "--n-epochs", type=int, default=5,
help="number of training epochs")
parser.add_argument("--l2norm", type=float, default=0,
help="l2 norm coef")
parser.add_argument("--relabel", default=False, action='store_true',
help="remove untouched nodes and relabel")
parser.add_argument("--fanout", type=str, default="5, 5",
help="Fan-out of neighbor sampling.")
parser.add_argument("--use-self-loop", default=False, action='store_true',
help="include self feature as a special relation")
fp = parser.add_mutually_exclusive_group(required=False)
fp.add_argument('--validation', dest='validation', action='store_true')
fp.add_argument('--testing', dest='validation', action='store_false')
parser.add_argument("--batch-size", type=int, default=1024,
help="Mini-batch size. ")
parser.add_argument("--eval-batch-size", type=int, default=1024,
help="Mini-batch size. ")
parser.add_argument("--num-workers", type=int, default=0,
help="Number of workers for dataloader.")
parser.add_argument("--low-mem", default=False, action='store_true',
help="Whether use low mem RelGraphCov")
parser.add_argument("--mix-cpu-gpu", default=False, action='store_true',
help="Whether store node embeddins in cpu")
parser.add_argument("--sparse-embedding", action='store_true',
help='Use sparse embedding for node embeddings.')
parser.add_argument('--node-feats', default=True, action='store_true',
help='Whether use node features')
parser.add_argument('--global-norm', default=False, action='store_true',
help='User global norm instead of per node type norm')
parser.add_argument('--layer-norm', default=False, action='store_true',
help='Use layer norm')
parser.set_defaults(validation=True)
args = parser.parse_args(args = [])
num_of_ntype = len(g.ntypes) # 1
num_rels = 149 # g.canonical_etypes = 1
num_classes = 2
train_idx = th.Tensor(train_idx).long()
val_idx = train_idx
test_idx = th.Tensor(test_idx).long()
labels = th.Tensor(labels).long()
6.5.训练和预测
args.gpu = '0'
devices = list(map(int, args.gpu.split(',')))
n_gpus = len(devices)
# cpu
if devices[0] == -1:
run(0, 0, args, ['cpu'],
(g, num_of_ntype, num_classes, num_rels, target_idx,
train_idx, val_idx, test_idx, labels))
# gpu
elif n_gpus == 1:
run(0, n_gpus, args, devices,
(g, node_feats, num_of_ntype, num_classes, num_rels, target_idx,
train_idx, val_idx, test_idx, labels), None, None)
# multi gpu
else:
procs = []
num_train_seeds = train_idx.shape[0]
tseeds_per_proc = num_train_seeds // n_gpus
for proc_id in range(n_gpus):
proc_train_seeds = train_idx[proc_id * tseeds_per_proc :
(proc_id + 1) * tseeds_per_proc \
if (proc_id + 1) * tseeds_per_proc < num_train_seeds \
else num_train_seeds]
p = mp.Process(target=run, args=(proc_id, n_gpus, args, devices,
(g, num_of_ntype, num_classes, num_rels, target_idx,
proc_train_seeds, val_idx, test_idx, labels)))
p.start()
procs.append(p)
for p in procs:
p.join()
6.6.加载模型并测试
model_ = th.load('./checkpoints/model_5.3_RGCN_nodefeature_mp.pkl')
dev_id = 0
queue=None
embed_layer = RelGraphEmbedLayer(dev_id,
g.number_of_nodes(),
node_tids,
num_of_ntype,
node_feats,
args.n_hidden,
sparse_emb=args.sparse_embedding)
test_sampler = NeighborSampler(g, target_idx, [None] * args.n_layers)
test_loader = DataLoader(dataset=test_idx.numpy(),
batch_size=args.eval_batch_size,
collate_fn=test_sampler.sample_blocks,
shuffle=False,
num_workers=args.num_workers)
model_.eval()
test_logits = []
test_seeds = []
with th.no_grad():
for sample_data in tqdm(test_loader):
th.cuda.empty_cache()
seeds, blocks = sample_data
if args.mix_cpu_gpu is False:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
else:
feats = embed_layer(blocks[0].srcdata[dgl.NID],
blocks[0].srcdata[dgl.NTYPE],
blocks[0].srcdata['type_id'],
node_feats)
logits = model_(blocks, feats)
test_logits.append(logits.cpu().detach())
test_seeds.append(seeds.cpu().detach())
test_logits = th.cat(test_logits)
test_seeds = th.cat(test_seeds)
if queue is not None:
queue.put((test_logits, test_seeds))
# if proc_id == 0:
if queue is not None:
test_logits = []
test_seeds = []
for i in range(n_gpus):
log = queue.get()
test_l, test_s = log
test_logits.append(test_l)
test_seeds.append(test_s)
test_logits = th.cat(test_logits)
test_seeds = th.cat(test_seeds)
test_loss = F.cross_entropy(test_logits, labels[test_seeds].cpu()).item()
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy = compute_acc(test_logits, labels[test_seeds])
print('Test: Neg Recall = {:.2f} | Neg Acc = {:.2f} | Pos Acc = {:.2f} | All Acc = {:.2f} | Loss = {:.4f}'.format(
test_neg_recall, test_neg_acc, test_pos_acc, test_accuracy, test_loss))
7.学习资料
DGL异构图教程:
https://docs.dgl.ai/tutorials/hetero/1_basics.html
DGL v0.4 更新笔记:
https://github.com/dmlc/dgl/releases
DGL-KE代码及使用说明:训练知识图谱嵌入(Knowledge Graph Embedding)专用包
https://github.com/dmlc/dgl/tree/master/apps/kg
DGL-Chem 模型库: 包括分子性质预测和分子结构生成等预训练模型。
https://docs.dgl.ai/api/python/model_zoo.html#chemistry
用于节点分类和链接预测等任务的RGCN和用于产品推荐的GCMC。
传统同构图(Homogeneous Graph)数据中只存在一种节点和边,因此在构建图神经网络时所有节点共享同样的模型参数并且拥有同样维度的特征空间。
而异构图(Heterogeneous Graph)中可以存在不只一种节点和边,因此允许不同类型的节点拥有不同维度的特征或属性。
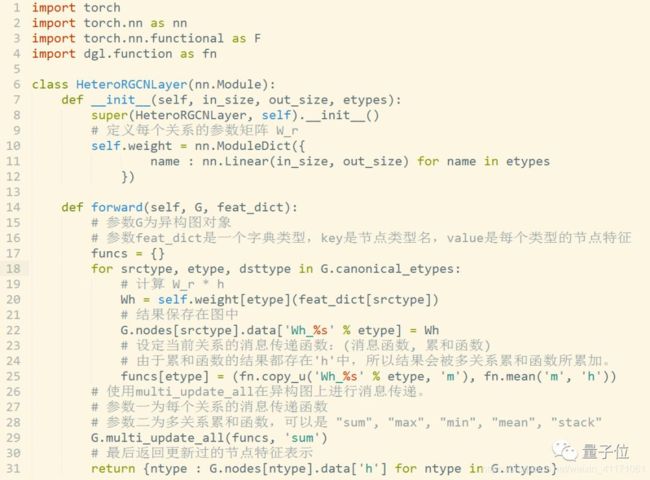
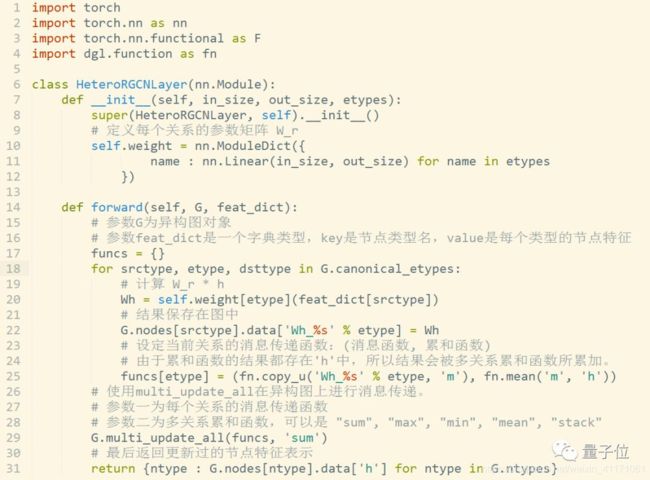
DGL 0.4中基于异构图的RGCN层实现代码