【机器学习的学习笔记】(持续更新)
Notes of studying Machine Learning
内容概览
- Data Preprocessing
- Supervised Learning
-
- Regression
- Terminologies
-
- Linear Regression
-
- Simple Linear Regression
- Multiple Linear Regression
- Polynomial Regression
- Support Vector Regression
- Decision Trees Regression
- Classification
-
- Binary Classifier
- Multi-class Classifier
- Learners in Classification Problems
-
- Lazy Learners
- Eager Learners
- Evaluating a Classification model
-
- 1. Log Loss or Cross-Entropy Loss
- 2. Confusion Matrix
- 3. AUC-ROC curve
- Linear Model
-
- Logistic Regression
-
- Binomial
- Multinomial
- Ordinal
- Non-linear Model
-
- Lazy Learners: K-Nearest Neighbours
- Eager Learners: Decision Trees
-
- Terminologies
- Random Forest [Overfitting]
- Eager Learners: Naïve Bayes
-
- Bernoulli, Multinomial and Gaussian Naive Bayes
- Assumption
- Support Vector Machines (SVM)
-
- Linear SVM
-
- Tune Parameter
- Non-linear SVM (Kernel SVM)
- Dimensionality Reduction
-
-
- Filters Methods
- Wrappers Methods
- Embedded Methods
- Lasso Regression/ L1 regularization [Reduce complexity]
- Ridge Regression/ L2 regularization [Reduce complexity]
-
- Feature Extractions
-
- Principal Component Analysis (Unsupervised Learning)
- Linear Discriminant Analysis
- Kernel PCA
- Quadratic Discriminant Analysis
- Unsupervised Learning
-
- Clustering
-
- Partitioning Clustering
- Density-Based Clustering
- Distribution Model-Based Clustering
- Hierarchical Clustering/ Agglomerative Hierarchical Clustering
- Fuzzy Clustering
- Association
-
- Apriori Algorithm
- Eclat Algorithm
- F-P Growth Algorithm
- Hyper Parameter Tuning
-
- Approach 1: Use train_test_split and manually tune parameters by trial and error
- Approach 2: Use K Fold Cross validation
- Approach 3: Use GridSearchCV
- Use RandomizedSearchCV to reduce number of iterations and with random combination of parameters.
- Different Models with Different Parameters
- K-Fold Cross Vaildation
- Bagging/ Ensemble Learning
- References
Data Preprocessing
机器学习的学习笔记。
Supervised Learning
Supervised learning is the types of machine learning in which machines are trained using well “labelled” training data, and on basis of that data, machines predict the output. The labelled data means some input data is already tagged with the correct output.
In the real-world, supervised learning can be used for Risk Assessment, Image Classification, Fraud Detection, Spam Filtering, etc.
Regression
- a relationship between the input variable and the output variable
- the prediction of continuous variables, such as
Weather Forecasting,Market Trends, etc.
Terminologies
-
Dependent Variable: The main factor in Regression analysis which we want to predict or understand is called the dependent variable. It is also called target variable.
-
Independent Variable: The factors which affect the dependent variables or which are used to predict the values of the dependent variables are called independent variable, also called as a predictor.
-
Outliers: Outlier is an observation which contains either very low value or very high value in comparison to other observed values. An outlier may hamper the result, so it should be avoided.
-
Multicollinearity: If the independent variables are highly correlated with each other than other variables, then such condition is called Multicollinearity. It should not be present in the dataset, because it creates problem while ranking the most affecting variable.
-
Underfitting and Overfitting: If our algorithm works well with the training dataset but not well with test dataset, then such problem is called Overfitting. And if our algorithm does not perform well even with training dataset, then such problem is called underfitting.
Linear Regression
Simple Linear Regression
The key point in Simple Linear Regression is that the dependent variable must be a continuous/real value. However, the independent variable can be measured on continuous or categorical values.
A single Independent/Predictor(x) variable is used to model the response variable (Y)
Y = b 0 + b 1 x Y = b_0+b_1x Y=b0+b1x
Y = dependent variables (target variables),
X = Independent variables (predictor variables),
a and b are the linear coefficients
Steps:
Data Pre-processing Steps
Fitting the MLR model to the training set
Predicting the result of the test set
Multiple Linear Regression
More than one predictor variable to predict the response variable
Y = b 0 + b 1 x + b 2 x + b 3 x + . . . . . + b n x Y = b_0+b_1x+ b_2x+ b_3x+.....+ b_nx Y=b0+b1x+b2x+b3x+.....+bnx
import dataset
import pandas as pd
import numpy as npdf = pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Supervised Learning/Linear Regression/homeprices.csv.xls')
area bedrooms age price
0 2600 3.0 20 550000
1 3000 4.0 15 565000
2 3200 NaN 18 610000
3 3600 3.0 30 595000
4 4000 5.0 8 760000
5 4100 6.0 8 810000
remove NA
import math
median_bedrooms = math.floor(df.bedrooms.median())
median_bedrooms
df.bedrooms = df.bedrooms.fillna(median_bedrooms) # clean your data, Data Preprocessing: Fill NA values with median value of a column
df
area bedrooms age price
0 2600 3.0 20 550000
1 3000 4.0 15 565000
2 3200 4.0 18 610000
3 3600 3.0 30 595000
4 4000 5.0 8 760000
5 4100 6.0 8 810000
import model to find the relationship between area, bedrooms, age, and the price
from sklearn import linear_model
reg = linear_model.LinearRegression()
reg.fit(df[['area','bedrooms','age']],df.price)
"\nfrom sklearn.linear_model import LinearRegression\nreg = LinearRegression()\nreg.fit(df[['area','bedrooms','age']], df.price)\n"
this is a 2D array of shape (n_targets, n_features),
see how different factor change the price
reg.coef_
reg.intercept_
reg.predict([[3000,3,40]]) # 3000 4.0 15 565000 Find price of home with 3000 sqr ft area, 3 bedrooms, 40 year old
# return
array([498408.25158031])
reg.predict([[2500,4,5]]) # 2600 3.0 20 550000 Find price of home with 2500 sqr ft area, 4 bedrooms, 5 year old
# return
array([578876.03748933])
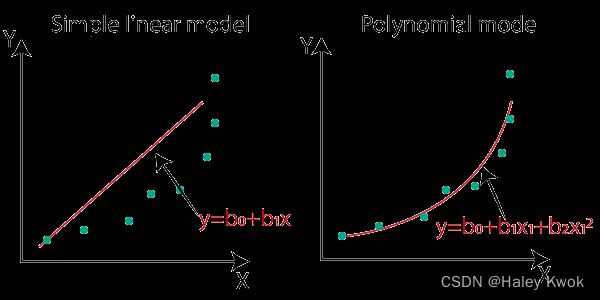
Polynomial Regression
Data points are arranged in a non-linear fashion, we need the Polynomial Regression model
Y = b 0 + b 1 x + b 2 x 2 + b 3 x 3 + . . . . . + b n x n Y = b_0+b_1x+ b_2x^2+ b_3x^3+.....+ b_nx^n Y=b0+b1x+b2x2+b3x3+.....+bnxn
Y is the predicted/target output, b0, b1,… bn are the regression coefficients. x is our independent/input variable.
Steps:
- Data Pre-processing
#importing libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#importing datasets
df = pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Supervised Learning/Linear Regression/Position_Salaries.csv')
df
# return
Position Level Salary
0 Business Analyst 1 45000
1 Junior Consultant 2 50000
2 Senior Consultant 3 60000
3 Manager 4 80000
4 Country Manager 5 110000
5 Region Manager 6 150000
6 Partner 7 200000
7 Senior Partner 8 300000
8 C-level 9 500000
9 CEO 10 1000000
#Extracting Independent and dependent Variable
X = df.iloc[:, 1:2].values # level
y = df.iloc[:, 2].values # salary
- Build a Linear Regression model and fit it to the dataset
#Fitting the Linear Regression to the dataset
from sklearn.linear_model import LinearRegression
lin_regs= LinearRegression()
lin_regs.fit(X,y)
- Build a Polynomial Regression model and fit it to the dataset
#Fitting the Polynomial regression to the dataset
from sklearn.preprocessing import PolynomialFeatures
poly_regs= PolynomialFeatures(degree= 2) # the polynomial degree depends on our choice
x_poly= poly_regs.fit_transform(X) # converting our feature matrix into polynomial feature matrix
lin_reg_2 =LinearRegression()
lin_reg_2.fit(x_poly, y)
- Visualize the result for Linear Regression and Polynomial Regression model
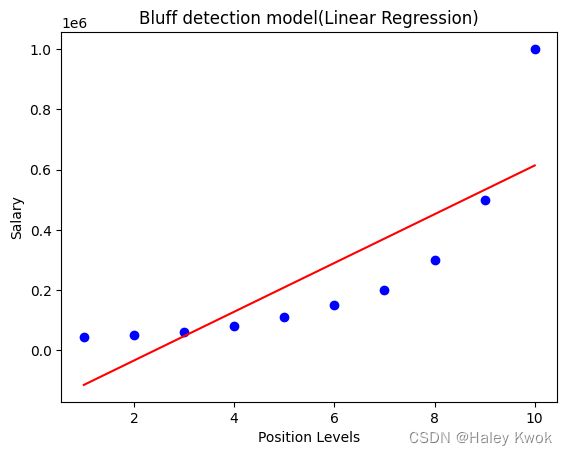
#Visulaizing the result for Linear Regression model
plt.scatter(X,y,color="blue")
plt.plot(X,lin_regs.predict(X), color="red")
plt.title("Bluff detection model(Linear Regression)")
plt.xlabel("Position Levels")
plt.ylabel("Salary")
plt.show()
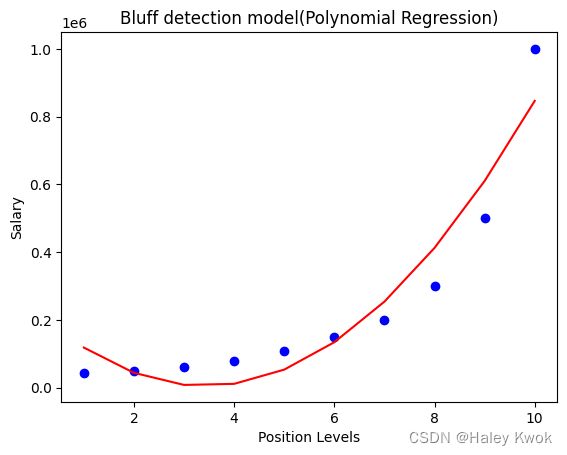
#Visulaizing the result for Polynomial Regression
plt.scatter(X,y,color="blue")
plt.plot(X, lin_reg_2.predict(poly_regs.fit_transform(X)), color="red")
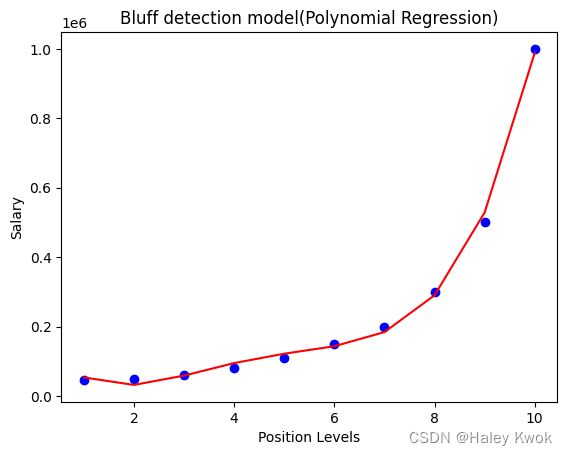
plt.title("Bluff detection model(Polynomial Regression)")
plt.xlabel("Position Levels")
plt.ylabel("Salary")
plt.show()
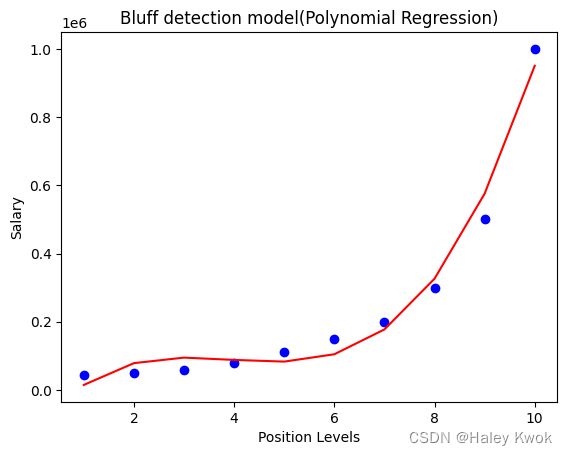
#Fitting the Polynomial regression to the dataset by degree=3
from sklearn.preprocessing import PolynomialFeatures
poly_regs= PolynomialFeatures(degree= 3) # the polynomial degree depends on our choice
x_poly= poly_regs.fit_transform(X) # converting our feature matrix into polynomial feature matrix
lin_reg_2 =LinearRegression()
lin_reg_2.fit(x_poly, y)
#Visulaizing the result for Polynomial Regression
plt.scatter(X,y,color="blue")
plt.plot(X, lin_reg_2.predict(poly_regs.fit_transform(X)), color="red")
plt.title("Bluff detection model(Polynomial Regression)")
plt.xlabel("Position Levels")
plt.ylabel("Salary")
plt.show()
when degree=4, the curve is smoother and more accurate
- Predicting the output
#Predicting the final result with the Linear Regression model:
lin_pred = lin_regs.predict([[6.5]])
print(lin_pred)
# return
[330378.78787879]
#Predicting the final result with the Polynomial Regression model:
poly_pred = lin_reg_2.predict(poly_regs.fit_transform([[6.5]]))
print(poly_pred)
# return
[158862.45265158]
Support Vector Regression
Decision Trees Regression
Classification
- the output variable is categorical
Binary Classifier
If the classification problem has only two possible outcomes, then it is called as Binary Classifier.
Examples: Yes-No, Male-Female, True-false, etc.
Multi-class Classifier
If a classification problem has more than two outcomes, then it is called as Multi-class Classifier.
Example: Classifications of types of crops, Classification of types of music.
Learners in Classification Problems
Lazy Learners
Lazy Learner firstly stores the training dataset and wait until it receives the test dataset. In Lazy learner case, classification is done on the basis of the most related data stored in the training dataset. It takes less time in training but more time for predictions.
Example: K-NN algorithm, Case-based reasoning
Eager Learners
Eager Learners develop a classification model based on a training dataset before receiving a test dataset. Opposite to Lazy learners, Eager Learner takes more time in learning, and less time in prediction.
Example: Decision Trees, Naïve Bayes, ANN.
Evaluating a Classification model
1. Log Loss or Cross-Entropy Loss
It is used for evaluating the performance of a classifier, whose output is a probability value between the 0 and 1.
For a good binary Classification model, the value of log loss should be near to 0.
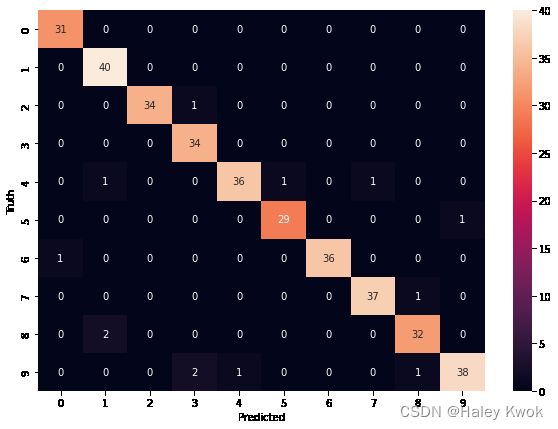
2. Confusion Matrix
- True Negative: Model has given prediction No, and the real or actual value was also No.
- True Positive: The model has predicted yes, and the actual value was also true.
- False Negative: The model has predicted no, but the actual value was Yes, it is also called as Type-II error.
- False Positive: The model has predicted Yes, but the actual value was No. It is also called a Type-I error.
y_predicted = model.predict(X_test)
from sklearn.metrics import confusion matrix
cm = confusion_matrix(y_test, y_predicted)
import seaborn as sn
plt.figure(figsize = (10,7))
sn.heatmap(cm, annot = True)
3. AUC-ROC curve
ROC curve stands for Receiver Operating Characteristics Curve and AUC stands for Area Under the Curve.
The ROC curve is plotted with TPR and FPR, where TPR (True Positive Rate) on Y-axis and FPR(False Positive Rate) on X-axis.
Linear Model
Logistic Regression
f ( x ) = 1 1 + e − x f(x)=\frac{1}{1+e^{-x}} f(x)=1+e−x1
uses sigmoid function or logistic function which is a complex cost function
f(x)= Output between the 0 and 1 value.
x= input to the function
e= base of natural logarithm.
Logistic Function (Sigmoid Function)
In logistic regression, we use the concept of the threshold value, which defines the probability of either 0 or 1. Such as values above the threshold value tends to 1, and a value below the threshold values tends to 0.
Assumptions
The dependent variable must be categorical in nature.
The independent variable should not have multi-collinearity.
Binomial
In binomial Logistic regression, there can be only two possible types of the dependent variables, such as 0 or 1, Pass or Fail, etc.
Steps:
- Data Pre-processing step
#Data Pre-procesing Step
# importing libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#importing datasets
data_set= pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Supervised Learning/Classification/Logistics Regression/User_Data.csv')
#Extracting Independent and dependent Variable
x= data_set.iloc[:, [2,3]].values
y= data_set.iloc[:, 4].values
# Splitting the dataset into training and test set.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test= train_test_split(x, y, test_size= 0.25, random_state=0)
#feature Scaling
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
x_train= st_x.fit_transform(x_train)
x_test= st_x.transform(x_test)
# return
User ID Gender Age EstimatedSalary Purchased
0 15624510 Male 19 19000 0
1 15810944 Male 35 20000 0
2 15668575 Female 26 43000 0
3 15603246 Female 27 57000 0
4 15804002 Male 19 76000 0
... ... ... ... ... ...
395 15691863 Female 46 41000 1
396 15706071 Male 51 23000 1
397 15654296 Female 50 20000 1
398 15755018 Male 36 33000 0
399 15594041 Female 49 36000 1
400 rows × 5 columns
- Fitting Logistic Regression to the Training set
#Fitting Logistic Regression to the training set
from sklearn.linear_model import LogisticRegression
classifier= LogisticRegression(random_state=0)
classifier.fit(x_train, y_train)
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='warn', n_jobs=None, penalty='l2',
random_state=0, solver='warn', tol=0.0001, verbose=0,
warm_start=False)
- Predicting the test result
#Predicting the test set result
y_pred= classifier.predict(x_test)
- Test accuracy of the result(Creation of Confusion matrix)
#Creating the Confusion matrix
from sklearn.metrics import confusion_matrix
cm= confusion_matrix(y_test, y_pred)
- Visualizing the test set result
#Visualizing the training set result
from matplotlib.colors import ListedColormap
x_set, y_set = x_train, y_train
x1, x2 = np.meshgrid(np.arange(start = x_set[:, 0].min() - 1, stop = x_set[:, 0].max() + 1, step =0.01),
np.arange(start = x_set[:, 1].min() - 1, stop = x_set[:, 1].max() + 1, step = 0.01))
plt.contourf(x1, x2, classifier.predict(np.array([x1.ravel(), x2.ravel()]).T).reshape(x1.shape),
alpha = 0.75, cmap = ListedColormap(('purple','green' )))
plt.xlim(x1.min(), x1.max())
plt.ylim(x2.min(), x2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(x_set[y_set == j, 0], x_set[y_set == j, 1],
c = ListedColormap(('purple', 'green'))(i), label = j)
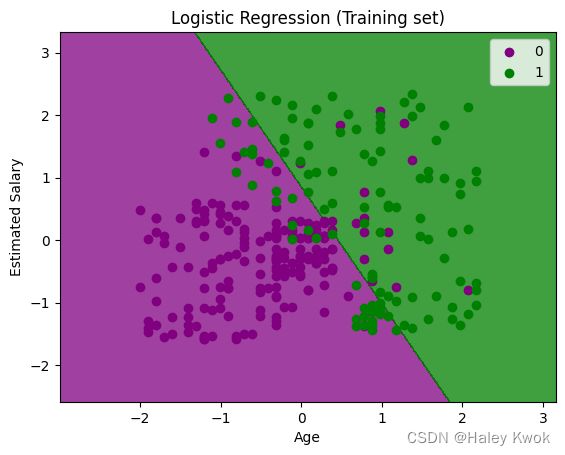
plt.title('Logistic Regression (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
Multinomial
In multinomial Logistic regression, there can be 3 or more possible unordered types of the dependent variable, such as “cat”, “dogs”, or “sheep”
from sklearn.datasets import load_digits
%matplotlib inline
import matplotlib.pyplot as plt
digits = load_digits()
dir(digits)
digits.data[0] #number 0
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(digits.data,digits.target, test_size=0.2)
model.fit(X_train, y_train)
model.score(X_test, y_test)
# return
0.9694444444444444
y_predicted = model.predict(X_test)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_predicted)
cm
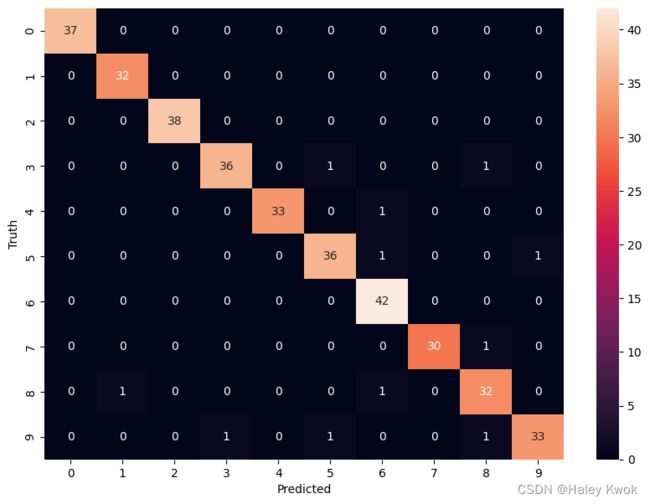
import seaborn as sn
plt.figure(figsize = (10,7))
sn.heatmap(cm, annot=True)
plt.xlabel('Predicted')
plt.ylabel('Truth')
Ordinal
In ordinal Logistic regression, there can be 3 or more possible ordered types of dependent variables, such as “low”, “Medium”, or “High”.
Non-linear Model
Lazy Learners: K-Nearest Neighbours
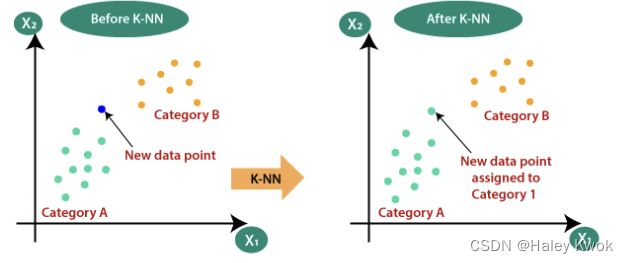
K-NN is a non-parametric algorithm, which means it does not make any assumption on underlying data.
KNN algorithm at the training phase just stores the dataset and when it gets new data, then it classifies that data into a category that is much similar to the new data.
Step-1: Select the number K of the neighbors
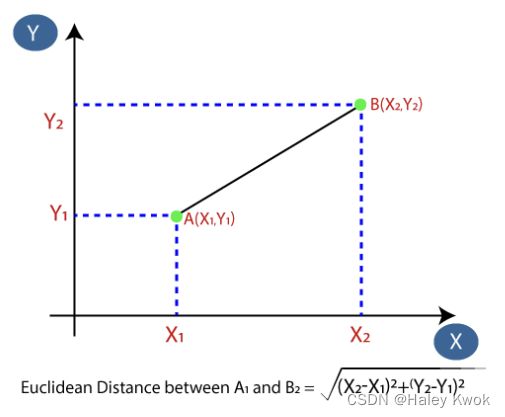
Step-2: Calculate the Euclidean distance of K number of neighbors
Step-3: Take the K nearest neighbors as per the calculated Euclidean distance.
Step-4: Among these k neighbors, count the number of the data points in each category.
Step-5: Assign the new data points to that category for which the number of the neighbor is maximum.
Step-6: Our model is ready.
- Data Pre-processing step
import pandas as pd
from sklearn.datasets import load_iris
iris = load_iris()
iris.feature_names
# return
['sepal length (cm)',
'sepal width (cm)',
'petal length (cm)',
'petal width (cm)']
iris.target_names
# return
array(['setosa', 'versicolor', 'virginica'], dtype=')
df = pd.DataFrame(iris.data,columns=iris.feature_names)
df.head()
# return
sepal length (cm) sepal width (cm) petal length (cm) petal width (cm)
0 5.1 3.5 1.4 0.2
1 4.9 3.0 1.4 0.2
2 4.7 3.2 1.3 0.2
3 4.6 3.1 1.5 0.2
4 5.0 3.6 1.4 0.2
df['target']=iris.target
df.head()
# return
sepal length (cm) sepal width (cm) petal length (cm) petal width (cm) target
0 5.1 3.5 1.4 0.2 0
1 4.9 3.0 1.4 0.2 0
2 4.7 3.2 1.3 0.2 0
3 4.6 3.1 1.5 0.2 0
4 5.0 3.6 1.4 0.2 0
df['flower_name'] =df.target.apply(lambda x: iris.target_names[x])
df.head()
# return
sepal length (cm) sepal width (cm) petal length (cm) petal width (cm) target flower_name
0 5.1 3.5 1.4 0.2 0 setosa
1 4.9 3.0 1.4 0.2 0 setosa
2 4.7 3.2 1.3 0.2 0 setosa
3 4.6 3.1 1.5 0.2 0 setosa
4 5.0 3.6 1.4 0.2 0 setosa
- Fitting the K-NN algorithm to the Training set
from sklearn.model_selection import train_test_split
X = df.drop(['target','flower_name'], axis='columns')
y = df.target
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=10) # Number of neighbors to use by default for
knn.fit(X_train, y_train)
# return
KNeighborsClassifier(n_neighbors=10)
knn.fit(X_test, y_test)
# return
KNeighborsClassifier(n_neighbors=10)
- Predicting the test result
knn.predict([[4.8,3.0,1.5,0.3]])
# return
/Users/haleyk/opt/anaconda3/lib/python3.8/site-packages/sklearn/base.py:450: UserWarning: X does not have valid feature names, but KNeighborsClassifier was fitted with feature names
warnings.warn(
array([0])
- Test accuracy of the result(Creation of Confusion matrix)
from sklearn.metrics import confusion_matrix
y_pred = knn.predict(X_test)
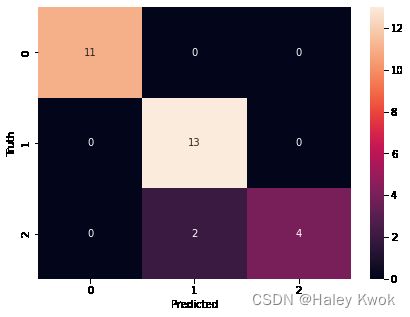
cm = confusion_matrix(y_test, y_pred)
cm
# return
array([[11, 0, 0],
[ 0, 13, 0],
[ 0, 2, 4]])
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sn
plt.figure(figsize=(7,5))
sn.heatmap(cm, annot=True)
plt.xlabel('Predicted')
plt.ylabel('Truth')
# 11+13+2+4 = 30 test set
# 2 is wrong
# there is only 2 wrong, therefore, the score is 0.933333
knn.score(X_test, y_test)
# return
0.9333333333333333
- Print classification report for precision, recall and f1-score for each classes
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# return
precision recall f1-score support
0 1.00 1.00 1.00 11
1 0.87 1.00 0.93 13
2 1.00 0.67 0.80 6
accuracy 0.93 30
macro avg 0.96 0.89 0.91 30
weighted avg 0.94 0.93 0.93 30
- Data Pre-processing step
# importing libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#importing datasets
data_set= pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Supervised Learning/Classification/Logistics Regression/User_Data.csv')
data_set
User ID Gender Age EstimatedSalary Purchased
0 15624510 Male 19 19000 0
1 15810944 Male 35 20000 0
2 15668575 Female 26 43000 0
3 15603246 Female 27 57000 0
4 15804002 Male 19 76000 0
... ... ... ... ... ...
395 15691863 Female 46 41000 1
396 15706071 Male 51 23000 1
397 15654296 Female 50 20000 1
398 15755018 Male 36 33000 0
399 15594041 Female 49 36000 1
400 rows × 5 columns
- Fitting the K-NN algorithm to the Training set
#Extracting Independent and dependent Variable
x= data_set.iloc[:, [2,3]].values
y= data_set.iloc[:, 4].values
# Splitting the dataset into training and test set.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test= train_test_split(x, y, test_size= 0.25, random_state=0)
#feature Scaling
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
x_train= st_x.fit_transform(x_train)
x_test= st_x.transform(x_test)
#Fitting K-NN classifier to the training set
from sklearn.neighbors import KNeighborsClassifier
classifier= KNeighborsClassifier(n_neighbors=5, metric='minkowski', p=2 )
classifier.fit(x_train, y_train)
- Predicting the test result
#Predicting the test set result
y_pred= classifier.predict(x_test)
- Test accuracy of the result(Creation of Confusion matrix)
#Creating the Confusion matrix
from sklearn.metrics import confusion_matrix
cm= confusion_matrix(y_test, y_pred)
- Visualizing the test set result
#Visulaizing the training set result
from matplotlib.colors import ListedColormap
x_set, y_set = x_train, y_train
x1, x2 = np.meshgrid(np.arange(start = x_set[:, 0].min() - 1, stop = x_set[:, 0].max() + 1, step =0.01),
np.arange(start = x_set[:, 1].min() - 1, stop = x_set[:, 1].max() + 1, step = 0.01))
plt.contourf(x1, x2, classifier.predict(np.array([x1.ravel(), x2.ravel()]).T).reshape(x1.shape),
alpha = 0.75, cmap = ListedColormap(('red','green' )))
plt.xlim(x1.min(), x1.max())
plt.ylim(x2.min(), x2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(x_set[y_set == j, 0], x_set[y_set == j, 1],
c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('K-NN Algorithm (Training set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
#Visualizing the test set result
from matplotlib.colors import ListedColormap
x_set, y_set = x_test, y_test
x1, x2 = np.meshgrid(np.arange(start = x_set[:, 0].min() - 1, stop = x_set[:, 0].max() + 1, step =0.01),
np.arange(start = x_set[:, 1].min() - 1, stop = x_set[:, 1].max() + 1, step = 0.01))
plt.contourf(x1, x2, classifier.predict(np.array([x1.ravel(), x2.ravel()]).T).reshape(x1.shape),
alpha = 0.75, cmap = ListedColormap(('red','green' )))
plt.xlim(x1.min(), x1.max())
plt.ylim(x2.min(), x2.max())
for i, j in enumerate(np.unique(y_set)):
plt.scatter(x_set[y_set == j, 0], x_set[y_set == j, 1],
c = ListedColormap(('red', 'green'))(i), label = j)
plt.title('K-NN algorithm(Test set)')
plt.xlabel('Age')
plt.ylabel('Estimated Salary')
plt.legend()
plt.show()
Eager Learners: Decision Trees
- solve problems for both categorical and numerical data
- builds a tree-like structure in which each internal node represents the “test” for an attribute, each branch represent the result of the test, and each leaf node represents the final decision or result.
- is constructed starting from the root node/parent node (dataset), which splits into left and right child nodes (subsets of dataset). These child nodes are further divided into their children node, and themselves become the parent node of those nodes.
Terminologies
Root Node: Root node is from where the decision tree starts. It represents the entire dataset, which further gets divided into two or more homogeneous sets.
Leaf Node: Leaf nodes are the final output node, and the tree cannot be segregated further after getting a leaf node.
Splitting: Splitting is the process of dividing the decision node/root node into sub-nodes according to the given conditions.
Branch/Sub Tree: A tree formed by splitting the tree.
Pruning: Pruning is the process of removing the unwanted branches from the tree.
Parent/Child node: The root node of the tree is called the parent node, and other nodes are called the child nodes.
Step-1: Begin the tree with the root node, says S, which contains the complete dataset.
Step-2: Find the best attribute in the dataset using Attribute Selection Measure (ASM).
- Information Gain
Information gain is the measurement of changes in entropy after the segmentation of a dataset based on an attribute.
Information Gain= Entropy(S)- [(Weighted Avg) *Entropy(each feature)
Entropy: Entropy is a metric to measure the impurity in a given attribute. It specifies randomness in data. Entropy can be calculated as:
Entropy(s)= -P(yes)log2 P(yes)- P(no) log2 P(no)
- Gini Index
Gini index is a measure of impurity or purity used while creating a decision tree in the CART(Classification and Regression Tree) algorithm.
An attribute with the low Gini index should be preferred as compared to the high Gini index.
G i n i I n d e x = 1 − ∑ j P j 2 Gini Index= 1- ∑ j_Pj_2 GiniIndex=1−∑jPj2
Step-3: Divide the S into subsets that contains possible values for the best attributes.
Step-4: Generate the decision tree node, which contains the best attribute.
Step-5: Recursively make new decision trees using the subsets of the dataset created in step -3. Continue this process until a stage is reached where you cannot further classify the nodes and called the final node as a leaf node.
-
Data Pre-Processing Step
-
Fitting a Decision-Tree algorithm to the Training set
#Fitting Decision Tree classifier to the training set
From sklearn.tree import DecisionTreeClassifier
classifier= DecisionTreeClassifier(criterion='entropy', random_state=0)
classifier.fit(x_train, y_train)
"criterion=‘entropy’: Criterion is used to measure the quality of split, which is calculated by information gain given by entropy.
- Predicting the test result
#Predicting the test set result
y_pred= classifier.predict(x_test)
- Test accuracy of the result (Creation of Confusion matrix)
#Creating the Confusion matrix
from sklearn.metrics import confusion_matrix
cm= confusion_matrix(y_test, y_pred)
# return
array([[62, 6],
[ 3, 29]])
In the above output image, we can see the confusion matrix, which has 6+3= 9 incorrect predictions and 62+29=91 correct predictions. Therefore, we can say that compared to other classification models, the Decision Tree classifier made a good prediction.
Another example:
import pandas as pd
df = pd.read_csv('/Users/haleyk/Desktop/Codebasics/ML/9_decision_tree/Exercise/titanic.csv')
df.head()
df.drop(['PassengerId','Name','SibSp','Parch','Ticket','Cabin','Embarked'],axis=1,inplace=True)
df.head()
df.Sex = df.Sex.map({'female':1, 'male':0})
df.Sex
df = df.fillna(df.Age.mean())
df
df.isnull().sum()
X = df.drop('Survived',axis='columns')
y = df.Survived
X
# return
Pclass Sex Age Fare
0 3 0 22.000000 7.2500
1 1 1 38.000000 71.2833
2 3 1 26.000000 7.9250
3 1 1 35.000000 53.1000
4 3 0 35.000000 8.0500
... ... ... ... ...
886 2 0 27.000000 13.0000
887 1 1 19.000000 30.0000
888 3 1 29.699118 23.4500
889 1 0 26.000000 30.0000
890 3 0 32.000000 7.7500
891 rows × 4 columns
y
# return
0 0
1 1
2 1
3 1
4 0
..
886 0
887 1
888 0
889 1
890 0
Name: Survived, Length: 891, dtype: int64
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size=0.2)
from sklearn import tree
model = tree.DecisionTreeClassifier()
model.fit(X_train,y_train)
# return
DecisionTreeClassifier()
model.score(X_test,y_test)
# return
0.8044692737430168
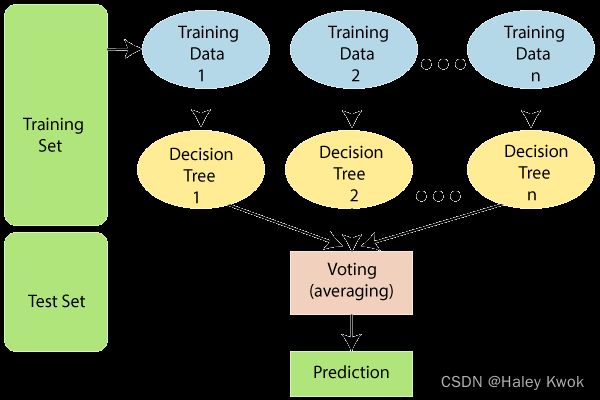
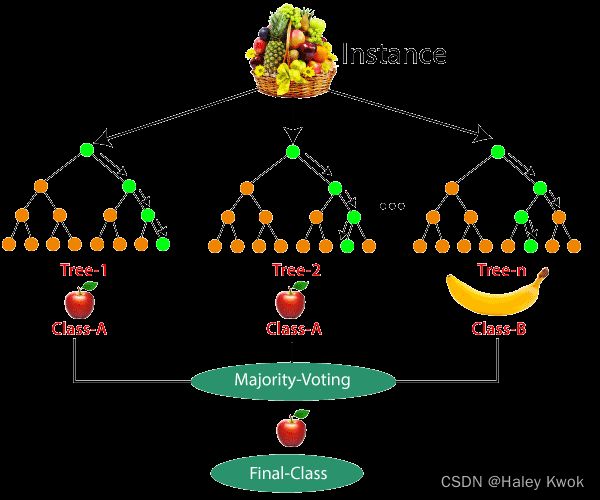
Random Forest [Overfitting]
- is one of the most powerful supervised learning algorithms which is capable of performing regression as well as classification tasks.
- is an ensemble learning method which combines multiple decision trees and predicts the final output based on the average of each tree output. The combined decision trees are called as base models.
- uses
BaggingorBootstrap Aggregationtechnique of ensemble learning in which aggregated decision tree runs in parallel and do not interact with each other.
g ( x ) = f 0 ( x ) + f 1 ( x ) + f 2 ( x ) + . . . g(x)= f_0(x)+ f_1(x)+ f_2(x)+... g(x)=f0(x)+f1(x)+f2(x)+...
Photo Source
Step-1: Select random K data points from the training set.
Step-2: Build the decision trees associated with the selected data points (Subsets).
Step-3: Choose the number N for decision trees that you want to build.
Step-4: Repeat Step 1 & 2.
Step-5: For new data points, find the predictions of each decision tree, and assign the new data points to the category that wins the majority votes.
Photo Source
import pandas as pd
from sklearn.datasets import load_digits
digits = load_digits()
dir(digits) # list of string
# return
['DESCR', 'data', 'feature_names', 'frame', 'images', 'target', 'target_names']
df[0:12]
X = df.drop('target',axis=1)
y = df.target
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=20) #The number of trees in the forest.
model.fit(X_train, y_train)
# return
RandomForestClassifier(n_estimators=20)
model.score(X_test, y_test)
# return
0.9638888888888889
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_predicted)
cm
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sn
plt.figure(figsize=(10,7))
sn.heatmap(cm, annot=True)
plt.xlabel('Predicted')
plt.ylabel('Truth')
# return
Text(69.0, 0.5, 'Truth')
Eager Learners: Naïve Bayes
Bernoulli, Multinomial and Gaussian Naive Bayes
- Multinomial Naïve Bayes consider a feature vector where a given term represents the number of times it appears or very often i.e. frequency.
- On the other hand, Bernoulli is a binary algorithm used when the feature is present or not.
- At last Gaussian is based on continuous distribution.
Naïve Bayes algorithm is a supervised learning algorithm, which is based on Bayes theorem and used for solving classification problems.
It is mainly used in text classification that includes a high-dimensional training dataset.
It is a probabilistic classifier, which means it predicts on the basis of the probability of an object.
Some popular examples of Naïve Bayes Algorithm are spam filtration, Sentimental analysis, and classifying articles.
- Naïve: It is called Naïve because it assumes that the occurrence of a certain feature is independent of the occurrence of other features. Such as if the fruit is identified on the bases of color, shape, and taste, then red, spherical, and sweet fruit is recognized as an apple. Hence each feature individually contributes to identify that it is an apple without depending on each other.
- Bayes: It is called Bayes because it depends on the principle of Bayes’ Theorem.
Bayes’ Theorem
P(A|B) = P(B|A)P(A) / P(B)
Assumption
Naive bayes theorm uses bayes theorm for conditional probability with a naive assumption that the features are not correlated to each other and tries to find conditional probability of target variable given the probabilities of features.
import pandas as pd
df = pd.read_csv('https://storage.googleapis.com/kagglesdsdata/competitions/3136/26502/train.csv?GoogleAccessId=web-data@kaggle-161607.iam.gserviceaccount.com&Expires=1646132661&Signature=PteVGgxccUCBs%2FsS7tCMNObj%2B6Svdf23ERl7SVnvVns2y4B7yPqW3mYYNQazGsbpEF6P0KUiKysl5ZTjIZEuNI%2FdduFeY77DWHdNVWZGDd5TlizWxOPd1E2rHcOyYpavsCr1OgtjLU8HpTCDsHhTb9kpzGoSK0Dzf2iFgh7HNWCsUYbHjCK9OAn6IBITg%2BZpn2KagdtPMFR3QWDeQV5kHlpsG9j8DaPdQGyJcZBC%2B8zuqz2UM6tknZp20fVDjtWR5ZvklfjzDpWGVtKxZfpFQlAv4f3udqF%2F%2F1j2zXFUcUXiD6tFdDR40rcNTL%2BVDJnUeNFHX4%2F7XbprMuToqMgpqQ%3D%3D&response-content-disposition=attachment%3B+filename%3Dtrain.csv')
df.head()
# return
PassengerId Survived Pclass Name Sex Age SibSp Parch Ticket Fare Cabin Embarked
0 1 0 3 Braund, Mr. Owen Harris male 22.0 1 0 A/5 21171 7.2500 NaN S
1 2 1 1 Cumings, Mrs. John Bradley (Florence Briggs Th... female 38.0 1 0 PC 17599 71.2833 C85 C
2 3 1 3 Heikkinen, Miss. Laina female 26.0 0 0 STON/O2. 3101282 7.9250 NaN S
3 4 1 1 Futrelle, Mrs. Jacques Heath (Lily May Peel) female 35.0 1 0 113803 53.1000 C123 S
4 5 0 3 Allen, Mr. William Henry male 35.0 0 0 373450 8.0500 NaN S
df.drop(['PassengerId','Name','SibSp','Parch','Ticket','Cabin','Embarked'],axis='columns',inplace=True)
df.head()
# return
Survived Pclass Sex Age Fare
0 0 3 male 22.0 7.2500
1 1 1 female 38.0 71.2833
2 1 3 female 26.0 7.9250
3 1 1 female 35.0 53.1000
4 0 3 male 35.0 8.0500
inputs = df.drop('Survived',axis='columns')
target = df.Survived
#inputs.Sex = inputs.Sex.map({'male': 1, 'female': 2})
dummies = pd.get_dummies(inputs.Sex)
dummies.head()
# return
female male
0 0 1
1 1 0
2 1 0
3 1 0
4 0 1
# merged
inputs = pd.concat([inputs,dummies],axis='columns')
inputs.head()
# return
Pclass Sex Age Fare female male
0 3 male 22.0 7.2500 0 1
1 1 female 38.0 71.2833 1 0
2 3 female 26.0 7.9250 1 0
3 1 female 35.0 53.1000 1 0
4 3 male 35.0 8.0500 0 1
inputs.drop(['Sex','male'],axis='columns',inplace=True)
inputs.head()
# return
Pclass Age Fare female
0 3 22.0 7.2500 0
1 1 38.0 71.2833 1
2 3 26.0 7.9250 1
3 1 35.0 53.1000 1
4 3 35.0 8.0500 0
inputs.columns[inputs.isna().any()] #detect missing data
# return
Index(['Age'], dtype='object')
inputs.Age[:10]
inputs.Age = inputs.Age.fillna(inputs.Age.mean()) # fill the NaN with mean value
inputs.head()
# return
Pclass Age Fare female
0 3 22.0 7.2500 0
1 1 38.0 71.2833 1
2 3 26.0 7.9250 1
3 1 35.0 53.1000 1
4 3 35.0 8.0500 0
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(inputs,target,test_size=0.3)
from sklearn.naive_bayes import GaussianNB
model = GaussianNB()
len(X_train)
# 623
len(X_test)
# 268
len(inputs)
# 891
model.fit(X_train,y_train)
model.score(X_test,y_test)
# return
0.7350746268656716
# Calculate the score using cross validation
from sklearn.model_selection import cross_val_score
cross_val_score(GaussianNB(),X_train, y_train, cv=5)
# return
array([0.784 , 0.808 , 0.784 , 0.76612903, 0.82258065])
Support Vector Machines (SVM)
Support Vector Machine is a supervised learning algorithm which can be used for regression as well as classification problems. So if we use it for regression problems, then it is termed as Support Vector Regression.
Photo Source
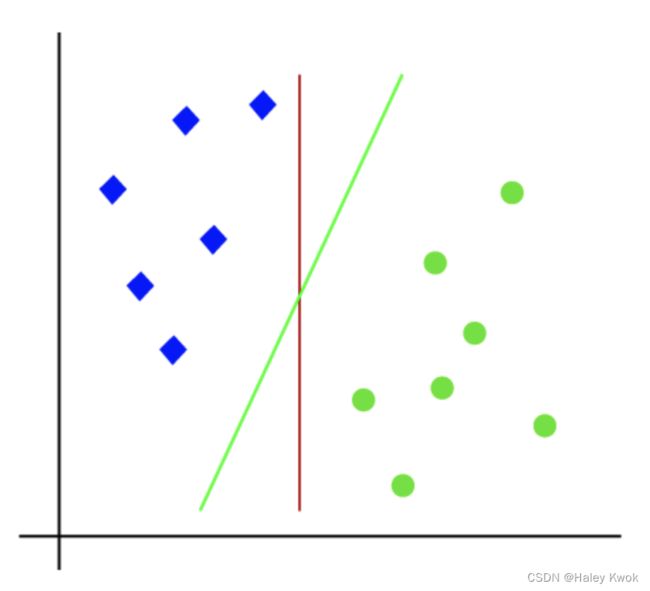
Linear SVM
Linear SVM is used for linearly separable data, which means if a dataset can be classified into two classes by using a single straight line, then such data is termed as linearly separable data, and classifier is used called as Linear SVM classifier.
Photo Source
#Data Pre-processing Step
# importing libraries
import numpy as nm
import matplotlib.pyplot as mtp
import pandas as pd
#importing datasets
data_set= pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Supervised Learning/Classification/Logistics Regression/User_Data.csv')
data_set
# return
User ID Gender Age EstimatedSalary Purchased
0 15624510 Male 19 19000 0
1 15810944 Male 35 20000 0
2 15668575 Female 26 43000 0
3 15603246 Female 27 57000 0
4 15804002 Male 19 76000 0
... ... ... ... ... ...
395 15691863 Female 46 41000 1
396 15706071 Male 51 23000 1
397 15654296 Female 50 20000 1
398 15755018 Male 36 33000 0
399 15594041 Female 49 36000 1
400 rows × 5 columns
#Extracting Independent and dependent Variable
x= data_set.iloc[:, [2,3]].values
y= data_set.iloc[:, 4].values
# Splitting the dataset into training and test set.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test= train_test_split(x, y, test_size= 0.25, random_state=0)
#feature Scaling
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
x_train= st_x.fit_transform(x_train)
x_test= st_x.transform(x_test)
from sklearn.svm import SVC # "Support vector classifier"
classifier = SVC(kernel='linear', random_state=0)
classifier.fit(x_train, y_train)
#Predicting the test set result
y_pred= classifier.predict(x_test)
#Creating the Confusion matrix
from sklearn.metrics import confusion_matrix
cm= confusion_matrix(y_test, y_pred)
cm
# return
array([[66, 2],
[ 8, 24]])
import pandas as pd
from sklearn.datasets import load_iris
iris = load_iris()
df['flower_name'] =df.target.apply(lambda x: iris.target_names[x])
df.head()
df['target'] = iris.target #0-49 setosa
df.head(
df[df.target==1].head() #50-99 versicolor
df[df.target==2].head() #100-149 virginica
df['flower_name'] =df.target.apply(lambda x: iris.target_names[x])
df.head()
from sklearn.model_selection import train_test_split
X = df.drop(['target','flower_name'], axis=1)
y = df.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
from sklearn.svm import SVC
model = SVC()
model.fit(X_train, y_train)
model.score(X_test, y_test)
# return
model.predict([[4.8,3.0,1.5,0.3]])
Tune Parameter
# Regularization (C)
model_C = SVC(C=1)
model_C.fit(X_train, y_train)
model_C.score(X_test, y_test)
model_C = SVC(C=10)
model_C.fit(X_train, y_train)
model_C.score(X_test, y_test)
# Gamma
model_g = SVC(gamma=10)
model_g.fit(X_train, y_train)
model_g.score(X_test, y_test)
# Kernel
model_linear_kernal = SVC(kernel='linear')
model_linear_kernal.fit(X_train, y_train)
model_linear_kernal.score(X_test, y_test)
Non-linear SVM (Kernel SVM)
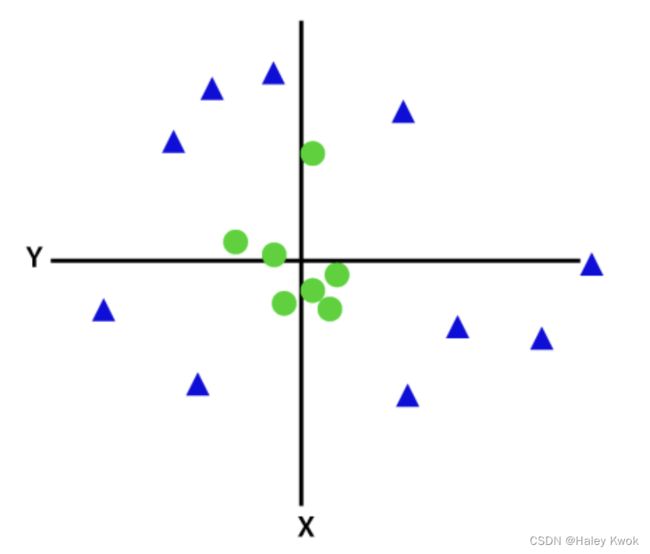
Non-Linear SVM is used for non-linearly separated data, which means if a dataset cannot be classified by using a straight line, then such data is termed as non-linear data and classifier used is called as Non-linear SVM classifier.
To separate these data points, we need to add one more dimension. For linear data, we have used two dimensions x and y, so for non-linear data, we will add a third dimension z.
z = x 2 + y 2 z=x^2 +y^2 z=x2+y2
#Data Pre-processing Step
# importing libraries
import numpy as nm
import matplotlib.pyplot as mtp
import pandas as pd
#importing datasets
data_set= pd.read_csv('user_data.csv')
#Extracting Independent and dependent Variable
x= data_set.iloc[:, [2,3]].values
y= data_set.iloc[:, 4].values
# Splitting the dataset into training and test set.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test= train_test_split(x, y, test_size= 0.25, random_state=0)
#feature Scaling
from sklearn.preprocessing import StandardScaler
st_x= StandardScaler()
x_train= st_x.fit_transform(x_train)
x_test= st_x.transform(x_test)
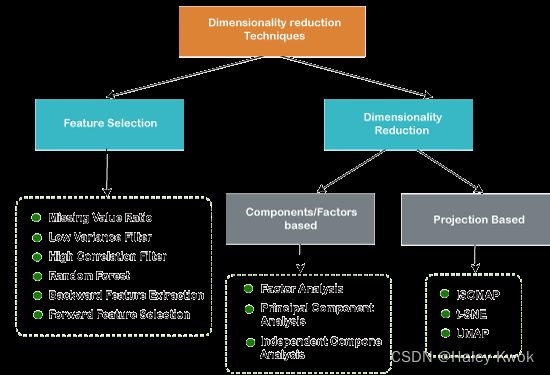
Dimensionality Reduction
Photo Source
Filters Methods
In this method, the dataset is filtered, and a subset that contains only the relevant features is taken. Some common techniques of filters method are:
Correlation
Chi-Square Test
ANOVA
Information Gain, etc.
Wrappers Methods
The performance decides whether to add those features or remove to increase the accuracy of the model. This method is more accurate than the filtering method but complex to work. Some common techniques of wrapper methods are:
Forward Selection
Backward Selection
Bi-directional Elimination
Embedded Methods
Embedded methods check the different training iterations of the machine learning model and evaluate the importance of each feature. Some common techniques of Embedded methods are:
Elastic Net
Lasso Regression/ L1 regularization [Reduce complexity]
- penalty term contains only the absolute weights
- can only shrink the slope to 0 because of taking absolute values
L o s s = E r r o r ( Y − Y ^ ) + λ ∑ 1 n ∣ w i ∣ Loss = Error(Y - \widehat{Y}) + \lambda \sum_1^n |w_i| Loss=Error(Y−Y )+λ1∑n∣wi∣
# import libraries
from sklearn.linear_model import Lasso
lasso_reg = Lasso(alpha=50, max_iter=100, tol=0.1)
lasso_reg.fit(train_X, train_y)
lasso_reg.score(test_X, test_y)
# return
0.6636
lasso_reg.score(train_X, train_y)
# return
0.6766
Ridge Regression/ L2 regularization [Reduce complexity]
- a general linear or polynomial regression will fail if there is high collinearity between the independent variables
- penalty term contains a square of weights
- shrink the slope near to 0
L o s s = E r r o r ( Y − Y ^ ) + λ ∑ 1 n w i 2 Loss = Error(Y - \widehat{Y}) + \lambda \sum_1^n w_i^{2} Loss=Error(Y−Y )+λ1∑nwi2
from sklearn.linear_model import Ridge
ridge_reg= Ridge(alpha=50, max_iter=100, tol=0.1)
ridge_reg.fit(train_X, train_y)
ridge_reg.score(test_X, test_y)
# return
0.6670848945194956
ridge_reg.score(train_X, train_y)
# return
0.6622376739684328
Feature Extractions
Principal Component Analysis (Unsupervised Learning)
Principal Component Analysis is a statistical process that converts the observations of correlated features into a set of linearly uncorrelated features with the help of orthogonal transformation. These new transformed features are called the Principal Components. It is one of the popular tools that is used for exploratory data analysis and predictive modeling.
from sklearn.decomposition import PCA
pca = PCA(0.95)
X_pca = pca.fit_transform(X)
X_pca.shape
# return
(1797, 29)
pca.explained_variance_ratio_
# return
array([0.14890594, 0.13618771, 0.11794594, 0.08409979, 0.05782415,
0.0491691 , 0.04315987, 0.03661373, 0.03353248, 0.03078806,
0.02372341, 0.02272697, 0.01821863, 0.01773855, 0.01467101,
0.01409716, 0.01318589, 0.01248138, 0.01017718, 0.00905617,
0.00889538, 0.00797123, 0.00767493, 0.00722904, 0.00695889,
0.00596081, 0.00575615, 0.00515158, 0.0048954 ])
pca.n_components_
# return
29
X_pca
# return
array([[ -1.25946645, 21.27488348, -9.46305462, ..., 3.67072108,
-0.9436689 , -1.13250195],
[ 7.9576113 , -20.76869896, 4.43950604, ..., 2.18261819,
-0.51022719, 2.31354911],
[ 6.99192297, -9.95598641, 2.95855808, ..., 4.22882114,
2.1576573 , 0.8379578 ],
...,
[ 10.8012837 , -6.96025223, 5.59955453, ..., -3.56866194,
1.82444444, 3.53885886],
[ -4.87210009, 12.42395362, -10.17086635, ..., 3.25330054,
0.95484174, -0.93895602],
[ -0.34438963, 6.36554919, 10.77370849, ..., -3.01636722,
1.29752723, 2.58810313]])
X_train_pca, X_test_pca, y_train, y_test = train_test_split(X_pca, y, test_size=0.2, random_state=30)
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(max_iter=1000)
model.fit(X_train_pca, y_train)
model.score(X_test_pca, y_test)
# return
0.9694444444444444
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X)
X_pca.shape
# return
(1797, 2)
X_pca
# return
array([[ -1.25946639, 21.27487891],
[ 7.95760922, -20.76869518],
[ 6.99192341, -9.95598163],
...,
[ 10.80128435, -6.96025523],
[ -4.87210315, 12.42395926],
[ -0.34438701, 6.36554335]])
pca.explained_variance_ratio_
# return: You can see that both combined retains 0.14+0.13=0.27 or 27% of important feature information
array([0.14890594, 0.13618771])
X_train_pca, X_test_pca, y_train, y_test = train_test_split(X_pca, y, test_size=0.2, random_state=30)
model = LogisticRegression(max_iter=1000)
model.fit(X_train_pca, y_train)
model.score(X_test_pca, y_test)
# return
0.6083333333333333
Linear Discriminant Analysis
Kernel PCA
Quadratic Discriminant Analysis
Unsupervised Learning
Unsupervised learning is a machine learning technique in which models are not supervised using training dataset. Instead, models itself find the hidden patterns and insights from the given data. It can be compared to learning which takes place in the human brain while learning new things.
The goal of unsupervised learning is to find the underlying structure of dataset, group that data according to similarities, and represent that dataset in a compressed format.
K-means clustering
KNN (k-nearest neighbors)
Anomaly detection
Neural Networks
Principle Component Analysis <>
Independent Component Analysis
Apriori algorithm
Singular value decomposition
Clustering
The clustering methods are broadly divided into Hard clustering (datapoint belongs to only one group) and Soft Clustering (data points can belong to another group also). But there are also other various approaches of Clustering exist.
Step-1: Select the number K to decide the number of clusters.
Step-2: Select random K points or centroids. (It can be other from the input dataset).
Step-3: Assign each data point to their closest centroid, which will form the predefined K clusters.
Step-4: Calculate the variance and place a new centroid of each cluster.
Step-5: Repeat the third steps, which means reassign each datapoint to the new closest centroid of each cluster.
Step-6: If any reassignment occurs, then go to step-4 else go to FINISH.
Step-7: The model is ready.
Elbow Method
WCSS stands for Within Cluster Sum of Squares, which defines the total variations within a cluster.
W C S S = ∑ P i i n C l u s t e r 1 d i s t a n c e ( P i C 1 ) 2 + ∑ P i i n C l u s t e r 2 d i s t a n c e P i C 2 2 + ∑ P i i n C l u s t e r 3 d i s t a n c e ( P i C 3 ) 2 WCSS= ∑P_i in Cluster1 distance(P_i C_1)^2 +∑P_i in Cluster2 distance P_i C_2^2+∑P_i in Cluster3 distance(P_i C_3)^2 WCSS=∑PiinCluster1distance(PiC1)2+∑PiinCluster2distancePiC22+∑PiinCluster3distance(PiC3)2
To measure the distance between data points and centroid, we can use any method such as Euclidean distance or Manhattan distance.
# importing libraries
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# Importing the dataset
dataset = pd.read_csv('/Users/haleyk/Documents/Python_Libraries_for_ML/Python_Libraries_for_ML/Unsupervised Learning/K-mean clustering/Mall_Customers.csv')
dataset
# return
CustomerID Gender Age Annual Income (k$) Spending Score (1-100)
0 1 Male 19 15 39
1 2 Male 21 15 81
2 3 Female 20 16 6
3 4 Female 23 16 77
4 5 Female 31 17 40
... ... ... ... ... ...
245 246 Male 30 297 69
246 247 Female 56 311 14
247 248 Male 29 313 90
248 249 Female 19 316 32
249 250 Female 31 325 86
250 rows × 5 columns
x = dataset.iloc[:, [3, 4]].values
#finding optimal number of clusters using the elbow method
from sklearn.cluster import KMeans
wcss_list= [] #Initializing the list for the values of WCSS
#Using for loop for iterations from 1 to 10.
for i in range(1, 11):
kmeans = KMeans(n_clusters=i, init='k-means++', random_state= 42)
kmeans.fit(x)
wcss_list.append(kmeans.inertia_)
plt.plot(range(1, 11), wcss_list)
plt.title('The Elobw Method Graph')
plt.xlabel('Number of clusters(k)')
plt.ylabel('wcss_list')
plt.show()
#training the K-means model on a dataset
kmeans = KMeans(n_clusters=5, init='k-means++', random_state= 42)
y_predict= kmeans.fit_predict(x)
#visulaizing the clusters
plt.scatter(x[y_predict == 0, 0], x[y_predict == 0, 1], s = 100, c = 'blue', label = 'Cluster 1') #for first cluster
plt.scatter(x[y_predict == 1, 0], x[y_predict == 1, 1], s = 100, c = 'green', label = 'Cluster 2') #for second cluster
plt.scatter(x[y_predict== 2, 0], x[y_predict == 2, 1], s = 100, c = 'red', label = 'Cluster 3') #for third cluster
plt.scatter(x[y_predict == 3, 0], x[y_predict == 3, 1], s = 100, c = 'cyan', label = 'Cluster 4') #for fourth cluster
plt.scatter(x[y_predict == 4, 0], x[y_predict == 4, 1], s = 100, c = 'magenta', label = 'Cluster 5') #for fifth cluster
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], s = 300, c = 'yellow', label = 'Centroid')
plt.title('Clusters of customers')
plt.xlabel('Annual Income (k$)')
plt.ylabel('Spending Score (1-100)')
plt.legend()
plt.show()
Below are the main clustering methods used in Machine learning:
Partitioning Clustering
It is a type of clustering that divides the data into non-hierarchical groups. It is also known as the centroid-based method. The most common example of partitioning clustering is the K-Means Clustering algorithm.
Density-Based Clustering
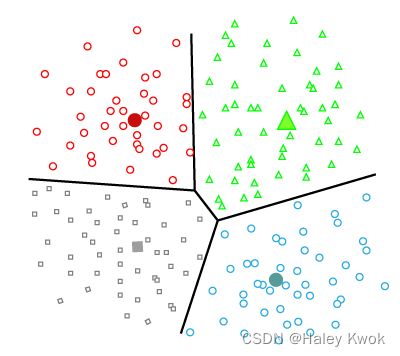
The density-based clustering method connects the highly-dense areas into clusters, and the arbitrarily shaped distributions are formed as long as the dense region can be connected. This algorithm does it by identifying different clusters in the dataset and connects the areas of high densities into clusters. The dense areas in data space are divided from each other by sparser areas.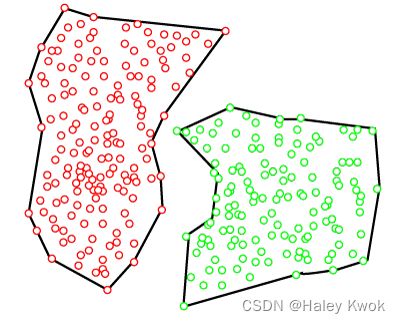
Distribution Model-Based Clustering
In the distribution model-based clustering method, the data is divided based on the probability of how a dataset belongs to a particular distribution. The grouping is done by assuming some distributions commonly Gaussian Distribution.
The example of this type is the Expectation-Maximization Clustering algorithm that uses Gaussian Mixture Models (GMM).

Hierarchical Clustering/ Agglomerative Hierarchical Clustering
In this technique, the dataset is divided into clusters to create a tree-like structure, which is also called a dendrogram. Hierarchical clustering is another unsupervised machine learning algorithm, which is used to group the unlabeled datasets into a cluster and also known as hierarchical cluster analysis or HCA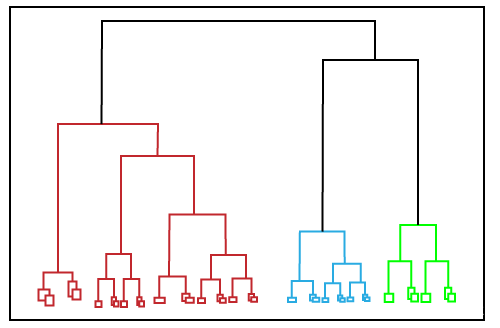
Fuzzy Clustering
Fuzzy clustering is a type of soft method in which a data object may belong to more than one group or cluster. Each dataset has a set of membership coefficients, which depend on the degree of membership to be in a cluster. Fuzzy C-means algorithm is the example of this type of clustering; it is sometimes also known as the Fuzzy k-means algorithm.
Association
Apriori Algorithm
This algorithm uses frequent datasets to generate association rules. It is designed to work on the databases that contain transactions. This algorithm uses a breadth-first search and Hash Tree to calculate the itemset efficiently.
It is mainly used for market basket analysis and helps to understand the products that can be bought together. It can also be used in the healthcare field to find drug reactions for patients.
Eclat Algorithm
Eclat algorithm stands for Equivalence Class Transformation. This algorithm uses a depth-first search technique to find frequent itemsets in a transaction database. It performs faster execution than Apriori Algorithm.
F-P Growth Algorithm
The F-P growth algorithm stands for Frequent Pattern, and it is the improved version of the Apriori Algorithm. It represents the database in the form of a tree structure that is known as a frequent pattern or tree. The purpose of this frequent tree is to extract the most frequent patterns.
Hyper Parameter Tuning
Approach 1: Use train_test_split and manually tune parameters by trial and error
from sklearn import svm, datasets
iris = datasets.load_iris()
import pandas as pd
df = pd.DataFrame(iris.data,columns=iris.feature_names)
df['flower'] = iris.target
df['flower'] = df['flower'].apply(lambda x: iris.target_names[x])
df[47:150]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(iris.data, iris.target, test_size=0.3)
model = svm.SVC(kernel='rbf',C=30,gamma='auto')
model.fit(X_train,y_train)
model.score(X_test, y_test)
# return
1
Approach 2: Use K Fold Cross validation
Manually try suppling models with different parameters to cross_val_score function with 5 fold cross validation
from sklearn.model_selection import cross_val_score
cross_val_score(svm.SVC(kernel='linear',C=10,gamma='auto'),iris.data, iris.target, cv=5)
cross_val_score(svm.SVC(kernel='rbf',C=10,gamma='auto'),iris.data, iris.target, cv=5)
cross_val_score(svm.SVC(kernel='linear',C=20,gamma='auto'),iris.data, iris.target, cv=5)
.....
Above approach is tiresome and very manual. We can use for loop as an alternative
import numpy as np
kernels = ['rbf', 'linear']
C = [1,10,20]
avg_scores = {}
for kval in kernels:
for cval in C:
cv_scores = cross_val_score(svm.SVC(kernel=kval,C=cval,gamma='auto'),iris.data, iris.target, cv=5)
avg_scores[kval + '_' + str(cval)] = np.average(cv_scores)
avg_scores
# return
{'rbf_1': 0.9800000000000001,
'rbf_10': 0.9800000000000001,
'rbf_20': 0.9666666666666668,
'linear_1': 0.9800000000000001,
'linear_10': 0.9733333333333334,
'linear_20': 0.9666666666666666}
From above results we can say that rbf with C=1 or 10 or linear with C=1 will give best performance
Approach 3: Use GridSearchCV
GridSearchCV does exactly same thing as for loop above but in a single line of code
from sklearn.model_selection import GridSearchCV
clf = GridSearchCV(svm.SVC(gamma='auto'), {
'C': [1,10,20], # parameter
'kernel': ['rbf','linear']
}, cv=5, return_train_score=False)
clf.fit(iris.data, iris.target)
clf.cv_results_
df = pd.DataFrame(clf.cv_results_)
df
df[['param_C','param_kernel','mean_test_score']]
clf.best_params_
clf.best_score_
Another Example:
Use RandomizedSearchCV to reduce number of iterations and with random combination of parameters.
This is useful when you have too many parameters to try and your training time is longer. It helps reduce the cost of computation
from sklearn.model_selection import RandomizedSearchCV
rs = RandomizedSearchCV(svm.SVC(gamma='auto'), {
'C': [1,10,20],
'kernel': ['rbf','linear']
},
cv=5,
return_train_score=False,
n_iter=2
)
rs.fit(iris.data, iris.target)
pd.DataFrame(rs.cv_results_)[['param_C','param_kernel','mean_test_score']]
# return
param_C param_kernel mean_test_score
0 1 linear 0.98
1 1 rbf 0.98
Different Models with Different Parameters
from sklearn import svm
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
model_params = {
'svm': {
'model': svm.SVC(gamma='auto'),
'params' : {
'C': [1,10,20],
'kernel': ['rbf','linear']
}
},
'random_forest': {
'model': RandomForestClassifier(),
'params' : {
'n_estimators': [1,5,10]
}
},
'logistic_regression' : {
'model': LogisticRegression(solver='liblinear',multi_class='auto'),
'params': {
'C': [1,5,10]
}
}
}
scores = []
for model_name, mp in model_params.items():
clf = GridSearchCV(mp['model'], mp['params'], cv=5, return_train_score=False)
clf.fit(iris.data, iris.target)
scores.append({
'model': model_name,
'best_score': clf.best_score_,
'best_params': clf.best_params_
})
df = pd.DataFrame(scores,columns=['model','best_score','best_params'])
df
# return
model best_score best_params
0 svm 0.980000 {'C': 1, 'kernel': 'rbf'}
1 random_forest 0.960000 {'n_estimators': 5}
2 logistic_regression 0.966667 {'C': 5}
Another Example:
from sklearn import datasets
digits = datasets.load_digits()
from sklearn import svm
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.naive_bayes import MultinomialNB
from sklearn.tree import DecisionTreeClassifier
model_params = {
'svm': {
'model': svm.SVC(gamma='auto'),
'params' : {
'C': [1,10,20],
'kernel': ['rbf','linear']
}
},
'random_forest': {
'model': RandomForestClassifier(),
'params' : {
'n_estimators': [1,5,10]
}
},
'logistic_regression' : {
'model': LogisticRegression(solver='liblinear',multi_class='auto'),
'params': {
'C': [1,5,10]
}
},
'naive_bayes_gaussian': {
'model': GaussianNB(),
'params': {}
},
'naive_bayes_multinomial': {
'model': MultinomialNB(),
'params': {}
},
'decision_tree': {
'model': DecisionTreeClassifier(),
'params': {
'criterion': ['gini','entropy'],
}
}
}
from sklearn.model_selection import GridSearchCV
import pandas as pd
scores = []
for model_name, mp in model_params.items():
clf = GridSearchCV(mp['model'], mp['params'], cv=5, return_train_score=False)
clf.fit(digits.data, digits.target)
scores.append({
'model': model_name,
'best_score': clf.best_score_,
'best_params': clf.best_params_
})
df = pd.DataFrame(scores,columns=['model','best_score','best_params'])
df
# return
model best_score best_params
0 svm 0.949360 {'C': 1, 'kernel': 'linear'}
1 random_forest 0.899833 {'n_estimators': 10}
2 logistic_regression 0.920979 {'C': 1}
3 naive_bayes_gaussian 0.806344 {}
4 naive_bayes_multinomial 0.871452 {}
5 decision_tree 0.817474 {'criterion': 'entropy'}
K-Fold Cross Vaildation
# option 1
# use all available data for training and testing on same dataset
# e.g., 100 math questions prepared, and a few questions to be tested
# option 2
# 70/100 for training and 30/100 for testing
# option 3
# K-fold cross vaildation
# 1000 samples 20/100 for testing and 80/100 for training x 5 sets
# take average
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
import numpy as np
from sklearn.datasets import load_digits
import matplotlib.pyplot as plt
digits = load_digits()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(digits.data,digits.target,test_size=0.3)
# Logistic Regression
lr = LogisticRegression(solver='liblinear',multi_class='ovr')
lr.fit(X_train, y_train)
lr.score(X_test, y_test)
# SVM
svm = SVC(gamma='auto')
svm.fit(X_train, y_train)
svm.score(X_test, y_test)
from sklearn.model_selection import cross_val_score
# cross_val_score uses stratifield kfold by default
# Logistic regression model performance using cross_val_score
cross_val_score(LogisticRegression(solver='liblinear',multi_class='ovr'), digits.data, digits.target,cv=3)
# svm model performance using cross_val_score
cross_val_score(SVC(gamma='auto'), digits.data, digits.target,cv=3)
# random forest performance using cross_val_score
cross_val_score(RandomForestClassifier(n_estimators=40),digits.data, digits.target,cv=3)
# Here we used cross_val_score to fine tune our random forest classifier and
# figured that having around 40 trees in random forest gives best result.
n_estimators=[5,20,30,40]
average_scores ={}
for ne in n_estimators:
cv_scores = cross_val_score(RandomForestClassifier(n_estimators=ne),digits.data, digits.target, cv=10)
average_scores['the average score of' +'_'+str(ne)+'_'+'is'] = np.average(cv_scores)
average_scores
Bagging/ Ensemble Learning
Ensemble learning is all about using multiple models to combine their prediction power to get better predictions that has low variance. Bagging and boosting are two popular techniques that allows us to tackle high variance issue. In this video we will learn about bagging with simple visual demonstration. We will also right python code in sklearn to use BaggingClassifier.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_scaled[:3]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, stratify=y, random_state=10)
from sklearn.model_selection import cross_val_score
from sklearn.tree import DecisionTreeClassifier
scores = cross_val_score(DecisionTreeClassifier(), X, y, cv=5)
scores
# return
0.7136575842458195
from sklearn.ensemble import BaggingClassifier
bag_model = BaggingClassifier(
base_estimator=DecisionTreeClassifier(),
n_estimators=100,
max_samples=0.8,
oob_score=True,
random_state=0
)
bag_model.fit(X_train, y_train)
bag_model.oob_score_
bag_model.score(X_test, y_test)
bag_model = BaggingClassifier(
base_estimator=DecisionTreeClassifier(),
n_estimators=100, # use 100 datasets, trial and error
max_samples=0.8, # 80% of my sample dataset
oob_score=True, # out of bag, since the data is random, then you use sth data that does not appear to test
random_state=0 # Controls the random resampling of the original dataset (sample wise and feature wise).
)
scores = cross_val_score(bag_model, X, y, cv=5)
scores
scores.mean()
# return
0.7578728461081402
# We can see some improvement in test score with bagging classifier as compared to a standalone classifier
from sklearn.ensemble import RandomForestClassifier
scores = cross_val_score(RandomForestClassifier(n_estimators=50), X, y, cv=5)
scores.mean()
# return
0.7669637551990494
from sklearn.svm import SVC
from sklearn.model_selection import cross_val_score
scores = cross_val_score(SVC(), X, y, cv=5)
scores.mean()
from sklearn.ensemble import BaggingClassifier
bag_model = BaggingClassifier(base_estimator=SVC(), n_estimators=100, max_samples=0.8, random_state=0)
scores = cross_val_score(bag_model, X, y, cv=5)
scores.mean()
References
Javapoint: Machine Learning
Scikit-learn