3DInfomax 化合物分子基于对比学习的向量表示及相似检索
参考:https://github.com/HannesStark/3DInfomax

1、3DInfomax 化合物分子基于对比学习的向量表示
首先把相关下载下来:
git clone https://github.com/HannesStark/3DInfomax.git
1)数据分子准备和yml文件创建
##数据加载 1万个smi分子,文件下载可参考:https://github.com/zilliztech/MolSearch/edit/master/script/test_1w.smi
mols2 = []
with open(r"test_1w.smi","r") as f:
ss = f.readlines()
# print(ss)
for i in ss:
mols2.append(i.strip().split()[0])
with open("smiles_1000.txt","w") as f:
for kk in mols2[:1000]:
f.write(kk+"\n")
simles_to_vecs.yml
smiles_txt_path: smiles_1000.txt
checkpoint: runs/PNA_qmugs_NTXentMultiplePositives_620000_123_25-08_09-19-52/best_checkpoint_35epochs.pt
2)项目根目录下创建个生成多个分子的向量表示的simles_to_vecs.py
import argparse
import concurrent.futures
import copy
import os
import re
from icecream import install
from ogb.lsc import DglPCQM4MDataset, PCQM4MEvaluator
from ogb.utils import smiles2graph
from commons.utils import seed_all, get_random_indices, TENSORBOARD_FUNCTIONS, move_to_device
from datasets.ZINC_dataset import ZINCDataset
from datasets.bace_geomol_feat import BACEGeomol
from datasets.bace_geomol_featurization_of_qm9 import BACEGeomolQM9Featurization
from datasets.bace_geomol_random_split import BACEGeomolRandom
from datasets.bbbp_geomol_feat import BBBPGeomol
from datasets.bbbp_geomol_featurization_of_qm9 import BBBPGeomolQM9Featurization
from datasets.bbbp_geomol_random_split import BBBPGeomolRandom
from datasets.esol_geomol_feat import ESOLGeomol
from datasets.esol_geomol_featurization_of_qm9 import ESOLGeomolQM9Featurization
from datasets.file_loader_drugs import FileLoaderDrugs
from datasets.file_loader_qm9 import FileLoaderQM9
from datasets.geom_drugs_dataset import GEOMDrugs
from datasets.geom_qm9_dataset import GEOMqm9
from datasets.geomol_geom_qm9_dataset import QM9GeomolFeatDataset
from datasets.lipo_geomol_feat import LIPOGeomol
from datasets.lipo_geomol_featurization_of_qm9 import LIPOGeomolQM9Featurization
from datasets.ogbg_dataset_extension import OGBGDatasetExtension
from datasets.qm9_dataset_geomol_conformers import QM9DatasetGeomolConformers
from datasets.qm9_dataset_rdkit_conformers import QM9DatasetRDKITConformers
from datasets.qm9_geomol_featurization import QM9GeomolFeaturization
from datasets.qmugs_dataset import QMugsDataset
from models.geomol_mpnn import GeomolGNNWrapper
from train import load_model
from trainer.byol_trainer import BYOLTrainer
from trainer.byol_wrapper import BYOLwrapper
import seaborn
from trainer.graphcl_trainer import GraphCLTrainer
from trainer.optimal_transport_trainer import OptimalTransportTrainer
from trainer.philosophy_trainer import PhilosophyTrainer
from trainer.self_supervised_ae_trainer import SelfSupervisedAETrainer
from trainer.self_supervised_alternating_trainer import SelfSupervisedAlternatingTrainer
from trainer.self_supervised_trainer import SelfSupervisedTrainer
import yaml
from datasets.custom_collate import * # do not remove
from models import * # do not remove
from torch.nn import * # do not remove
from torch.optim import * # do not remove
from commons.losses import * # do not remove
from torch.optim.lr_scheduler import * # do not remove
from datasets.samplers import * # do not remove
from datasets.qm9_dataset import QM9Dataset
from torch.utils.data import DataLoader, Subset
from trainer.metrics import QM9DenormalizedL1, QM9DenormalizedL2, \
QM9SingleTargetDenormalizedL1, Rsquared, NegativeSimilarity, MeanPredictorLoss, \
PositiveSimilarity, ContrastiveAccuracy, TrueNegativeRate, TruePositiveRate, Alignment, Uniformity, \
BatchVariance, DimensionCovariance, MAE, PositiveSimilarityMultiplePositivesSeparate2d, \
NegativeSimilarityMultiplePositivesSeparate2d, OGBEvaluator, PearsonR, PositiveProb, NegativeProb, \
Conformer2DVariance, Conformer3DVariance, PCQM4MEvaluatorWrapper
from trainer.trainer import Trainer
import gc
# turn on for debugging C code like Segmentation Faults
import faulthandler
faulthandler.enable()
install()
seaborn.set_theme()
from datasets.inference_dataset import InferenceDataset
def parse_arguments():
p = argparse.ArgumentParser()
p.add_argument('--config', type=argparse.FileType(mode='r'), default='configs_clean/fingerprint_inference.yml')
p.add_argument('--experiment_name', type=str, help='name that will be added to the runs folder output')
p.add_argument('--logdir', type=str, default='runs', help='tensorboard logdirectory')
p.add_argument('--num_epochs', type=int, default=2500, help='number of times to iterate through all samples')
p.add_argument('--batch_size', type=int, default=1024, help='samples that will be processed in parallel')
p.add_argument('--patience', type=int, default=20, help='stop training after no improvement in this many epochs')
p.add_argument('--minimum_epochs', type=int, default=0, help='minimum numer of epochs to run')
p.add_argument('--dataset', type=str, default='qm9', help='[qm9, zinc, drugs, geom_qm9, molhiv]')
p.add_argument('--num_train', type=int, default=-1, help='n samples of the model samples to use for train')
p.add_argument('--seed', type=int, default=123, help='seed for reproducibility')
p.add_argument('--num_val', type=int, default=None, help='n samples of the model samples to use for validation')
p.add_argument('--multithreaded_seeds', type=list, default=[],
help='if this is non empty, multiple threads will be started, training the same model but with the different seeds')
p.add_argument('--seed_data', type=int, default=123, help='if you want to use a different seed for the datasplit')
p.add_argument('--loss_func', type=str, default='MSELoss', help='Class name of torch.nn like [MSELoss, L1Loss]')
p.add_argument('--loss_params', type=dict, default={}, help='parameters with keywords of the chosen loss function')
p.add_argument('--critic_loss', type=str, default='MSELoss', help='Class name of torch.nn like [MSELoss, L1Loss]')
p.add_argument('--critic_loss_params', type=dict, default={},
help='parameters with keywords of the chosen loss function')
p.add_argument('--optimizer', type=str, default='Adam', help='Class name of torch.optim like [Adam, SGD, AdamW]')
p.add_argument('--optimizer_params', type=dict, help='parameters with keywords of the chosen optimizer like lr')
p.add_argument('--lr_scheduler', type=str,
help='Class name of torch.optim.lr_scheduler like [CosineAnnealingLR, ExponentialLR, LambdaLR]')
p.add_argument('--lr_scheduler_params', type=dict, help='parameters with keywords of the chosen lr_scheduler')
p.add_argument('--scheduler_step_per_batch', default=True, type=bool,
help='step every batch if true step every epoch otherwise')
p.add_argument('--log_iterations', type=int, default=-1,
help='log every log_iterations iterations (-1 for only logging after each epoch)')
p.add_argument('--expensive_log_iterations', type=int, default=100,
help='frequency with which to do expensive logging operations')
p.add_argument('--eval_per_epochs', type=int, default=0,
help='frequency with which to do run the function run_eval_per_epoch that can do some expensive calculations on the val set or sth like that. If this is zero, then the function will never be called')
p.add_argument('--linear_probing_samples', type=int, default=500,
help='number of samples to use for linear probing in the run_eval_per_epoch function of the self supervised trainer')
p.add_argument('--num_conformers', type=int, default=3,
help='number of conformers to use if we are using multiple conformers on the 3d side')
p.add_argument('--metrics', default=[], help='tensorboard metrics [mae, mae_denormalized, qm9_properties ...]')
p.add_argument('--main_metric', default='mae_denormalized', help='for early stopping etc.')
p.add_argument('--main_metric_goal', type=str, default='min', help='controls early stopping. [max, min]')
p.add_argument('--val_per_batch', type=bool, default=True,
help='run evaluation every batch and then average over the eval results. When running the molhiv benchmark for example, this needs to be Fale because we need to evaluate on all val data at once since the metric is rocauc')
p.add_argument('--tensorboard_functions', default=[], help='choices of the TENSORBOARD_FUNCTIONS in utils')
p.add_argument('--checkpoint', type=str, help='path to directory that contains a checkpoint to continue training')
p.add_argument('--pretrain_checkpoint', type=str, help='Specify path to finetune from a pretrained checkpoint')
p.add_argument('--transfer_layers', default=[],
help='strings contained in the keys of the weights that are transferred')
p.add_argument('--frozen_layers', default=[],
help='strings contained in the keys of the weights that are transferred')
p.add_argument('--exclude_from_transfer', default=[],
help='parameters that usually should not be transferred like batchnorm params')
p.add_argument('--transferred_lr', type=float, default=None, help='set to use a different LR for transfer layers')
p.add_argument('--num_epochs_local_only', type=int, default=1,
help='when training with OptimalTransportTrainer, this specifies for how many epochs only the local predictions will get a loss')
p.add_argument('--required_data', default=[],
help='what will be included in a batch like [dgl_graph, targets, dgl_graph3d]')
p.add_argument('--collate_function', default='graph_collate', help='the collate function to use for DataLoader')
p.add_argument('--collate_params', type=dict, default={},
help='parameters with keywords of the chosen collate function')
p.add_argument('--use_e_features', default=True, type=bool, help='ignore edge features if set to False')
p.add_argument('--targets', default=[], help='properties that should be predicted')
p.add_argument('--device', type=str, default='cuda', help='What device to train on: cuda or cpu')
p.add_argument('--dist_embedding', type=bool, default=False, help='add dist embedding to complete graphs edges')
p.add_argument('--num_radial', type=int, default=6, help='number of frequencies for distance embedding')
p.add_argument('--models_to_save', type=list, default=[],
help='specify after which epochs to remember the best model')
p.add_argument('--model_type', type=str, default='MPNN', help='Classname of one of the models in the models dir')
p.add_argument('--model_parameters', type=dict, help='dictionary of model parameters')
p.add_argument('--model3d_type', type=str, default=None, help='Classname of one of the models in the models dir')
p.add_argument('--model3d_parameters', type=dict, help='dictionary of model parameters')
p.add_argument('--critic_type', type=str, default=None, help='Classname of one of the models in the models dir')
p.add_argument('--critic_parameters', type=dict, help='dictionary of model parameters')
p.add_argument('--trainer', type=str, default='contrastive', help='[contrastive, byol, alternating, philosophy]')
p.add_argument('--train_sampler', type=str, default=None, help='any of pytorchs samplers or a custom sampler')
p.add_argument('--eval_on_test', type=bool, default=True, help='runs evaluation on test set if true')
p.add_argument('--force_random_split', type=bool, default=False, help='use random split for ogb')
p.add_argument('--reuse_pre_train_data', type=bool, default=False, help='use all data instead of ignoring that used during pre-training')
p.add_argument('--transfer_3d', type=bool, default=False, help='set true to load the 3d network instead of the 2d network')
p.add_argument('--smiles_txt_path', type=str, default='dataset/inference_smiles.txt', help='')
return p.parse_args()
def inference(args):
seed_all(args.seed)
device = torch.device("cuda:0" if torch.cuda.is_available() and args.device == 'cuda' else "cpu")
metrics_dict = {'rsquared': Rsquared(),
'mae': MAE(),
'pearsonr': PearsonR(),
'ogbg-molhiv': OGBEvaluator(d_name='ogbg-molhiv', metric='rocauc'),
'ogbg-molpcba': OGBEvaluator(d_name='ogbg-molpcba', metric='ap'),
'ogbg-molbace': OGBEvaluator(d_name='ogbg-molbace', metric='rocauc'),
'ogbg-molbbbp': OGBEvaluator(d_name='ogbg-molbbbp', metric='rocauc'),
'ogbg-molclintox': OGBEvaluator(d_name='ogbg-molclintox', metric='rocauc'),
'ogbg-moltoxcast': OGBEvaluator(d_name='ogbg-moltoxcast', metric='rocauc'),
'ogbg-moltox21': OGBEvaluator(d_name='ogbg-moltox21', metric='rocauc'),
'ogbg-mollipo': OGBEvaluator(d_name='ogbg-mollipo', metric='rmse'),
'ogbg-molmuv': OGBEvaluator(d_name='ogbg-molmuv', metric='ap'),
'ogbg-molsider': OGBEvaluator(d_name='ogbg-molsider', metric='rocauc'),
'ogbg-molfreesolv': OGBEvaluator(d_name='ogbg-molfreesolv', metric='rmse'),
'ogbg-molesol': OGBEvaluator(d_name='ogbg-molesol', metric='rmse'),
'pcqm4m': PCQM4MEvaluatorWrapper(),
'conformer_3d_variance': Conformer3DVariance(),
'conformer_2d_variance': Conformer2DVariance(),
'positive_similarity': PositiveSimilarity(),
'positive_similarity_multiple_positives_separate2d': PositiveSimilarityMultiplePositivesSeparate2d(),
'positive_prob': PositiveProb(),
'negative_prob': NegativeProb(),
'negative_similarity': NegativeSimilarity(),
'negative_similarity_multiple_positives_separate2d': NegativeSimilarityMultiplePositivesSeparate2d(),
'contrastive_accuracy': ContrastiveAccuracy(threshold=0.5009),
'true_negative_rate': TrueNegativeRate(threshold=0.5009),
'true_positive_rate': TruePositiveRate(threshold=0.5009),
'mean_predictor_loss': MeanPredictorLoss(globals()[args.loss_func](**args.loss_params)),
'uniformity': Uniformity(t=2),
'alignment': Alignment(alpha=2),
'batch_variance': BatchVariance(),
'dimension_covariance': DimensionCovariance()
}
# import tables
# import numpy as np
# filename = 'sim_vecs.h5'
# ROW_SIZE = 256
# # NUM_COLUMNS = 200
# f = tables.open_file(filename, mode='w')
# atom = tables.Float64Atom()
# array_c = f.create_earray(f.root, 'data', atom, (0, ROW_SIZE))
test_data = InferenceDataset(device=device, smiles_txt_path=args.smiles_txt_path)
print('num_smiles: ', len(test_data))
with open(args.smiles_txt_path) as file:
lines = file.readlines()
smiles_list = [line.rstrip() for line in lines]
print(args)
model, _,_ = load_model(args, data=test_data, device=device)
print('trainable params in model: ', sum(p.numel() for p in model.parameters() if p.requires_grad), '\n')
checkpoint = torch.load(args.checkpoint, map_location=device)
model.load_state_dict(checkpoint['model_state_dict'])
test_loader = DataLoader(test_data, batch_size=5, collate_fn=graph_only_collate)
# print(iter(test_loader).next())
fingerprints_list = []
with torch.no_grad():
for i, batch in enumerate(test_loader):
print("batch:",batch)
resultss = model(batch)
print("resultss:",resultss)
# new_resultss = resultss.detach().numpy()
# new_resultss = resultss.squeeze().detach().numpy()
# print("new_resultss:",new_resultss)
for ii in resultss:
fingerprints_list.append(ii.detach().numpy())
# fingerprints_list+=[ii]
# array_c.append(ii)
gc.collect()
# f.close()
print(fingerprints_list[:5])
# path = os.path.join('dataset', f'fingerprints.pt')
# print(f'Saving predictions to {path}')
# torch.save({'fingerprints': torch.cat(fingerprints_list, dim=0)}, path)
# all_vecs = torch.cat(fingerprints_list).squeeze().detach().numpy()
np.save(r"sims_1000.npy", smiles_list)
np.save(r"sim_vecs.npy", fingerprints_list)
# print(all_vecs.shape,all_vecs)
def get_arguments():
args = parse_arguments()
if args.config:
config_dict = yaml.load(args.config, Loader=yaml.FullLoader)
arg_dict = args.__dict__
for key, value in config_dict.items():
if isinstance(value, list):
for v in value:
arg_dict[key].append(v)
else:
arg_dict[key] = value
else:
config_dict = {}
# overwrite args with args from checkpoint except for the args that were contained in the config file
arg_dict = args.__dict__
with open(os.path.join(os.path.dirname(args.checkpoint), 'train_arguments.yaml'), 'r') as arg_file:
checkpoint_dict = yaml.load(arg_file, Loader=yaml.FullLoader)
for key, value in checkpoint_dict.items():
if key not in config_dict.keys():
if isinstance(value, list):
for v in value:
arg_dict[key].append(v)
else:
arg_dict[key] = value
return args
if __name__ == '__main__':
args = get_arguments()
inference(args)
3)运行上面py文件,保存向量表示npy格式
python simles_to_vecs.py --config=configs_clean/simles_to_vecs.yml
2、相似检索
1)单个分子向量表示计算
可以直接运行该文件
import torch
import dgl
import torch_geometric
from ogb.utils.features import atom_to_feature_vector, bond_to_feature_vector, get_atom_feature_dims, \
get_bond_feature_dims
from rdkit import Chem
from rdkit.Chem.rdmolops import GetAdjacencyMatrix
from torch.utils.data import Dataset
import numpy as np
import pandas as pd
from tqdm import tqdm
import torch.nn.functional as F
from scipy.constants import physical_constants
from commons.spherical_encoding import dist_emb
hartree2eV = physical_constants['hartree-electron volt relationship'][0]
##稍微改写了,去掉读写文件
class InferenceDataset(Dataset):
def __init__(self, smiles_txt, device='cuda:0', transform=None, **kwargs):
# with open(smiles_txt_path) as file:
# lines = file.readlines()
# smiles_list = [line.rstrip() for line in lines]
smiles_list =[smiles_txt]
atom_slices = [0]
edge_slices = [0]
all_atom_features = []
all_edge_features = []
edge_indices = [] # edges of each molecule in coo format
total_atoms = 0
total_edges = 0
n_atoms_list = []
for mol_idx, smiles in tqdm(enumerate(smiles_list)):
# get the molecule using the smiles representation from the csv file
mol = Chem.MolFromSmiles(smiles)
# add hydrogen bonds to molecule because they are not in the smiles representation
mol = Chem.AddHs(mol)
n_atoms = mol.GetNumAtoms()
atom_features_list = []
for atom in mol.GetAtoms():
atom_features_list.append(atom_to_feature_vector(atom))
all_atom_features.append(torch.tensor(atom_features_list, dtype=torch.long))
edges_list = []
edge_features_list = []
for bond in mol.GetBonds():
i = bond.GetBeginAtomIdx()
j = bond.GetEndAtomIdx()
edge_feature = bond_to_feature_vector(bond)
# add edges in both directions
edges_list.append((i, j))
edge_features_list.append(edge_feature)
edges_list.append((j, i))
edge_features_list.append(edge_feature)
# Graph connectivity in COO format with shape [2, num_edges]
edge_index = torch.tensor(edges_list, dtype=torch.long).T
edge_features = torch.tensor(edge_features_list, dtype=torch.long)
edge_indices.append(edge_index)
all_edge_features.append(edge_features)
total_edges += len(edges_list)
total_atoms += n_atoms
edge_slices.append(total_edges)
atom_slices.append(total_atoms)
n_atoms_list.append(n_atoms)
self.n_atoms = torch.tensor(n_atoms_list)
self.atom_slices = torch.tensor(atom_slices, dtype=torch.long)
self.edge_slices = torch.tensor(edge_slices, dtype=torch.long)
self.edge_indices = torch.cat(edge_indices, dim=1)
self.all_atom_features = torch.cat(all_atom_features, dim=0)
self.all_edge_features = torch.cat(all_edge_features, dim=0)
def __len__(self):
return len(self.atom_slices) - 1
def __getitem__(self, idx):
e_start = self.edge_slices[idx]
e_end = self.edge_slices[idx + 1]
start = self.atom_slices[idx]
n_atoms = self.n_atoms[idx]
edge_indices = self.edge_indices[:, e_start: e_end]
g = dgl.graph((edge_indices[0], edge_indices[1]), num_nodes=n_atoms)
g.ndata['feat'] = self.all_atom_features[start: start + n_atoms]
g.edata['feat'] = self.all_edge_features[e_start: e_end]
return g
from train import load_model
from torch.utils.data import DataLoader
from datasets.custom_collate import graph_only_collate
import argparse
p = argparse.ArgumentParser()
args = p.parse_args()
## 填写单个化合物的simles
simles_mol = "CC(=O)Nc1ccc(cc1)C1NN=C(S1)c1ccccc1"
checkpoint ='runs/PNA_qmugs_NTXentMultiplePositives_620000_123_25-08_09-19-52/best_checkpoint_35epochs.pt'
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
test_data = InferenceDataset(device=device, smiles_txt=simles_mol)
print('num_smiles: ', len(test_data))
args.model_type = "PNA"
args.model_parameters={'aggregators': ['mean', 'max', 'min', 'std'], 'batch_norm_momentum': 0.93, 'dropout': 0.0, 'hidden_dim': 200, 'last_batch_norm': True, 'mid_batch_norm': True, 'posttrans_layers': 1, 'pretrans_layers': 2, 'propagation_depth': 7, 'readout_aggregators':
['min', 'max', 'mean'], 'readout_batchnorm': True, 'readout_hidden_dim': 200, 'readout_layers': 2, 'residual': True, 'scalers': ['identity', 'amplification', 'attenuation'], 'target_dim': 256}
args.pretrain_checkpoint=None
model, _,_ = load_model(args, data=test_data, device=device)
print('trainable params in model: ', sum(p.numel() for p in model.parameters() if p.requires_grad), '\n')
checkpoint = torch.load(checkpoint, map_location=device)
model.eval()
model.load_state_dict(checkpoint['model_state_dict'])
test_loader = DataLoader(test_data, batch_size=1, collate_fn=graph_only_collate)
simles_data = iter(test_loader).next()
print("simles_data:",simles_data)
resultss = model(simles_data)
print("resultss:",resultss)
new_resultss = resultss.squeeze().detach().numpy()
print(new_resultss.shape,new_resultss)
np.save(r"single_vec.npy", new_resultss)
2)相似检索计算
import numpy as np
import torch
## 加载计算的全部分子库向量
all_vecs = np.load(r"sim_vecs.npy", allow_pickle=True)
all_vecs_ = torch.Tensor([(torch.from_numpy(item) / torch.from_numpy(item).norm(dim=-1, keepdim=True)).cpu().numpy() for item in all_vecs])
sims_1000 = np.load(r"sims_1000.npy") ##这是全部分子库的simles格式
## 加载单个分子用来检索的
single_vec = np.load(r"single_vec.npy", allow_pickle=True)
single_vec_ = torch.Tensor((torch.from_numpy(single_vec) / torch.from_numpy(single_vec).norm(dim=-1, keepdim=True)).cpu().numpy()).unsqueeze(0)
## 检索
similarities = (all_vecs_ @ single_vec_.T).squeeze(1)
print(similarities)
best_photo_idx = np.argsort(similarities.numpy())[::-1]
simle_ids = [sims_1000[i] for i in best_photo_idx[:10]] ##取前10
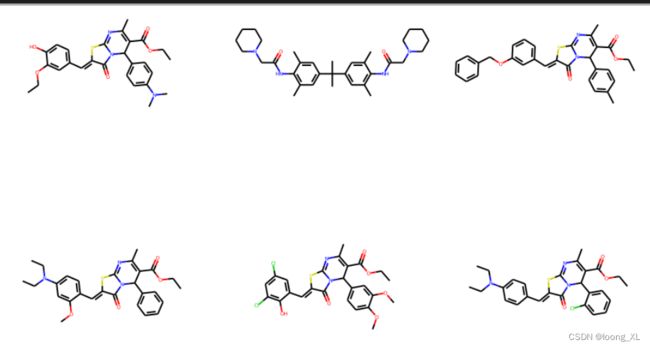
3)展示对比:
from rdkit.Chem import AllChem, Draw
from rdkit import Chem
Chem.MolFromSmiles("CC(=O)OC=N") ##单个分子
Draw.MolsToGridImage([ Chem.MolFromSmiles(i) for i in simle_ids],subImgSize=(300,300), molsPerRow=3) ## 前10相似
Chem.MolFromSmiles("CC(=O)Nc1ccc(cc1)C1NN=C(S1)c1ccccc1") ##单个分子
Draw.MolsToGridImage([ Chem.MolFromSmiles(i) for i in simle_ids],subImgSize=(300,300), molsPerRow=3) ## 前10相似