Python 11种数据降维算法快速实现,手把手教学
文章目录
- 环境要求
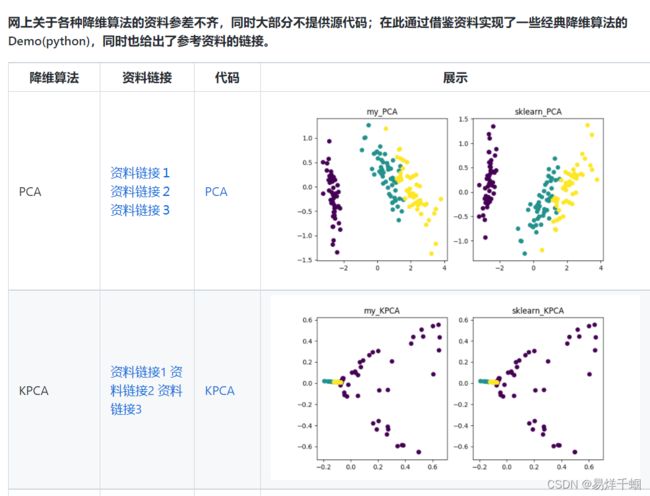
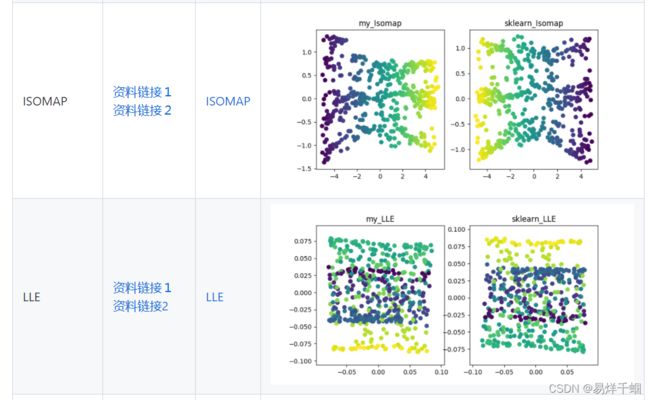
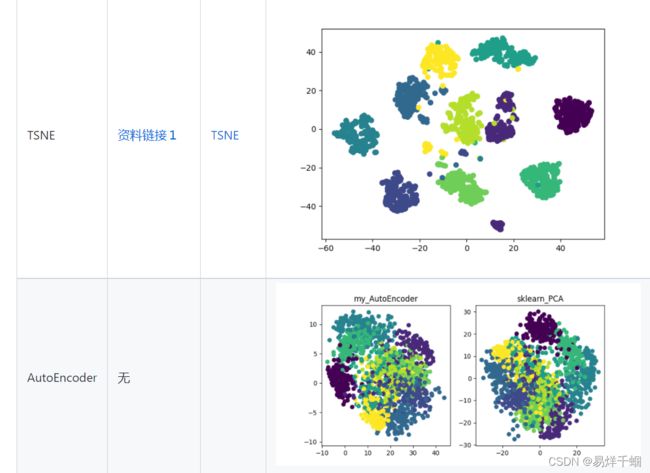
- 有哪些降维算法
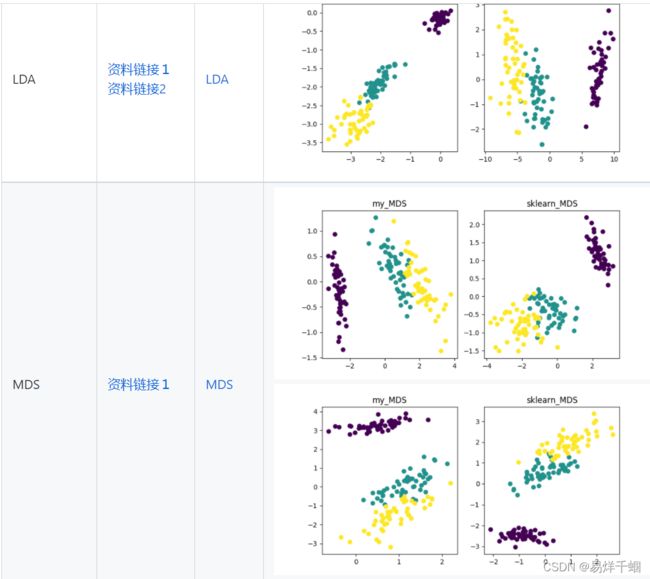
- LDA
- MDS
听说点进蝈仔帖子的都喜欢点赞加关注~~
老规矩,地址送上。
Github 项目地址:
https://github.com/heucoder/dimensionality_reduction_alo_codes
项目作者简介
Heucoder,目前是哈尔滨工业大学计算机技术在读硕士生,主要活跃于互联网领域,知乎昵称为「超爱学习」,其 github 主页地址为:https://github.com/heucoder。
环境要求
环境: python3.6 ubuntu18.04(windows10) 需要的库: numpy sklearn tensorflow matplotlib
每一个代码都可以单独运行,但是只是作为一个demo,仅供学习使用
其中AutoEncoder只是使用AutoEncoder简单的实现了一个PCA降维算法,自编码器涉及到了深度学习领域,其本身就是一个非常大领域
LE算法的鲁棒性极差,对近邻的选择和数据分布十分敏感
2019.6.20添加了LPP算法,但是效果没有论文上那么好,有点迷,后续需要修改
有哪些降维算法
LDA
线性判别分析(Linear Discriminant Analysis,LDA)是一种可作为特征抽取的技术
LDA可以提高数据分析过程中的计算效率,对于未能正则化的模型,可以降低维度灾难带来的过拟合。
话不多说,代码送上
#coding:utf-8
import numpy as np
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.datasets import load_iris
import matplotlib.pyplot as plt
def lda(data, target, n_dim):
'''
:param data: (n_samples, n_features)
:param target: data class
:param n_dim: target dimension
:return: (n_samples, n_dims)
'''
clusters = np.unique(target)
if n_dim > len(clusters)-1:
print("K is too much")
print("please input again")
exit(0)
#within_class scatter matrix
Sw = np.zeros((data.shape[1],data.shape[1]))
for i in clusters:
datai = data[target == i]
datai = datai-datai.mean(0)
Swi = np.mat(datai).T*np.mat(datai)
Sw += Swi
#between_class scatter matrix
SB = np.zeros((data.shape[1],data.shape[1]))
u = data.mean(0) #所有样本的平均值
for i in clusters:
Ni = data[target == i].shape[0]
ui = data[target == i].mean(0) #某个类别的平均值
SBi = Ni*np.mat(ui - u).T*np.mat(ui - u)
SB += SBi
S = np.linalg.inv(Sw)*SB
eigVals,eigVects = np.linalg.eig(S) #求特征值,特征向量
eigValInd = np.argsort(eigVals)
eigValInd = eigValInd[:(-n_dim-1):-1]
w = eigVects[:,eigValInd]
data_ndim = np.dot(data, w)
return data_ndim
if __name__ == '__main__':
iris = load_iris()
X = iris.data
Y = iris.target
data_1 = lda(X, Y, 2)
data_2 = LinearDiscriminantAnalysis(n_components=2).fit_transform(X, Y)
plt.figure(figsize=(8,4))
plt.subplot(121)
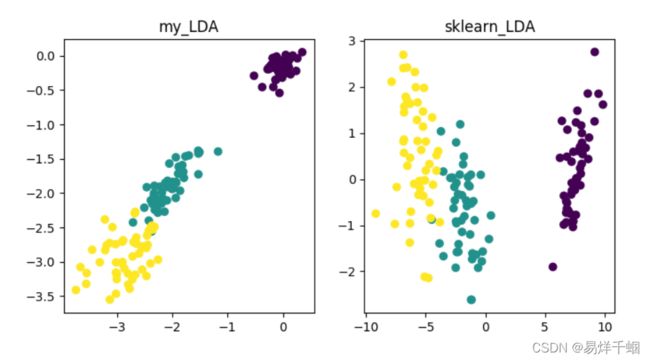
plt.title("my_LDA")
plt.scatter(data_1[:, 0], data_1[:, 1], c = Y)
plt.subplot(122)
plt.title("sklearn_LDA")
plt.scatter(data_2[:, 0], data_2[:, 1], c = Y)
plt.savefig("LDA.png")
plt.show()
MDS
非常传统的降维的方法,以距离为标准,将高维坐标中的点投影到低维坐标中,保持彼此之间的相对距离变化最小,更新的方法是T-SNE,基于分布概率变化最小进行投影。
# coding:utf-8
import numpy as np
from sklearn.datasets import load_iris
from sklearn.manifold import MDS
import matplotlib.pyplot as plt
def cal_pairwise_dist(x):
'''计算pairwise 距离, x是matrix
(a-b)^2 = a^2 + b^2 - 2*a*b
'''
sum_x = np.sum(np.square(x), 1)
dist = np.add(np.add(-2 * np.dot(x, x.T), sum_x).T, sum_x)
#返回任意两个点之间距离的平方
return dist
def my_mds(data, n_dims):
'''
:param data: (n_samples, n_features)
:param n_dims: target n_dims
:return: (n_samples, n_dims)
'''
n, d = data.shape
dist = cal_pairwise_dist(data)
dist[dist < 0 ] = 0
T1 = np.ones((n,n))*np.sum(dist)/n**2
T2 = np.sum(dist, axis = 1, keepdims=True)/n
T3 = np.sum(dist, axis = 0, keepdims=True)/n
B = -(T1 - T2 - T3 + dist)/2
eig_val, eig_vector = np.linalg.eig(B)
index_ = np.argsort(-eig_val)[:n_dims]
picked_eig_val = eig_val[index_].real
picked_eig_vector = eig_vector[:, index_]
# print(picked_eig_vector.shape, picked_eig_val.shape)
return picked_eig_vector*picked_eig_val**(0.5)
if __name__ == '__main__':
iris = load_iris()
data = iris.data
Y = iris.target
data_1 = my_mds(data, 2)
data_2 = MDS(n_components=2).fit_transform(data)
plt.figure(figsize=(8, 4))
plt.subplot(121)
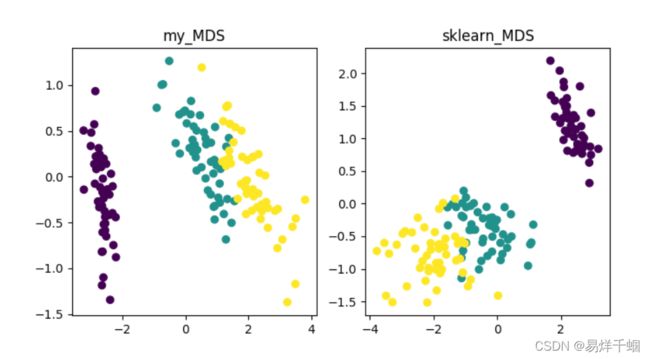
plt.title("my_MDS")
plt.scatter(data_1[:, 0], data_1[:, 1], c=Y)
plt.subplot(122)
plt.title("sklearn_MDS")
plt.scatter(data_2[:, 0], data_2[:, 1], c=Y)
plt.savefig("MDS_1.png")
plt.show()