R语言医学数据分析实战(二)数据框的操作
文章目录
- 一、用基本包处理数据框
-
-
- 1)查看数据框里的内容
- 2)选取数据框的子集
- 3)将数据框按照某个变量的值排序
- 4)查看和删除重复数据
- 5)在数据框中添加和删除变量
- 6)把数据框添加到搜索路径
-
- 二、使用dplyr包处理数据框
-
-
- 1)使用slice()和filter()筛选行
- 2)使用arrange()筛选行
- 3)使用select()选择列
- 4)使用mutate()添加新变量
- 5)使用summarise()计算统计量
- 6)使用group_by()拆分数据框
- 7)使用传递符‘%>%’组合多个操作
-
- 三、数据框的合并
-
-
- 1)纵向合并rbind()
- 2)横向合并cbind()
- 3)按照某个共有变量合并
-
- 四、数据框的长宽格式转换
- 五、缺失值的处理
-
-
- 1)识别缺失值
- 2)探索数据框里的缺失值
- 3)填充缺失值
-
- 六、处理大型数据集的策略
-
-
- 1)清理工作空间
- 2)模拟一个大型数据集
- 3)剔除不需要的变量
- 4)选取数据集的一个随机样本
-
一、用基本包处理数据框
导入Familydata数据:
> rm(list=ls()) #清空工作空间
> #install.packages("epiDisplay")
> library(epiDisplay)
> data(Familydata)
> ls() #查看工作空间
[1] "Familydata"
1)查看数据框里的内容
Familydata
head(Familydata)
tail(Familydata)
names(Familydata) #列出数据框中所有变量的名字
str(Familydata) #查看数据框结构
attributes(Familydata) #显示数据框属性的全部信息
#添加两个标签
attr(Familydata,"var.labels")[1]<-"ID num"
attr(Familydata,"var.labels")[6]<-"Gender"
attributes(Familydata)$var.labels
des(Familydata) #str()简化版
Anthropometric and financial data of a hypothetical family
No. of observations = 11
Variable Class Description
1 code character ID num
2 age integer Age(yr)
3 ht integer Ht(cm.)
4 wt integer Wt(kg.)
5 money integer Pocket money(B.)
6 sex factor Gender
2)选取数据框的子集
> Familydata[,3]
[1] 120 172 163 158 153 148 160 163 170 155 167
> Familydata$ht
[1] 120 172 163 158 153 148 160 163 170 155 167
> Familydata[1:3,c(3,4,6)]
ht wt sex
1 120 22 F
2 172 52 M
3 163 71 M
> subset(Familydata,sex=="F") #选取子集
code age ht wt money sex
1 K 6 120 22 5 F
4 I 18 158 51 200 F
5 C 69 153 51 300 F
6 B 72 148 60 500 F
7 G 46 160 50 500 F
8 H 42 163 55 600 F
10 F 47 155 53 2000 F
> subset(Familydata,sex=="F",select = c(ht,wt))
ht wt
1 120 22
4 158 51
5 153 51
6 148 60
7 160 50
8 163 55
10 155 53
> sample.rows<-sample(1:nrow(Familydata),size = 3,replace = FALSE) #sample():随机抽样
> sample.rows
[1] 10 7 4
> Familydata[sample.rows,]
code age ht wt money sex
10 F 47 155 53 2000 F
7 G 46 160 50 500 F
4 I 18 158 51 200 F
3)将数据框按照某个变量的值排序
> Familydata[order(Familydata$age),] #从小到大
code age ht wt money sex
1 K 6 120 22 5 F
2 J 16 172 52 50 M
4 I 18 158 51 200 F
8 H 42 163 55 600 F
7 G 46 160 50 500 F
10 F 47 155 53 2000 F
11 E 49 167 64 5000 M
9 D 58 170 67 2000 M
5 C 69 153 51 300 F
6 B 72 148 60 500 F
3 A 80 163 71 100 M
> Familydata[order(Familydata$age,decreasing = TRUE),] #从大到小
code age ht wt money sex
3 A 80 163 71 100 M
6 B 72 148 60 500 F
5 C 69 153 51 300 F
9 D 58 170 67 2000 M
11 E 49 167 64 5000 M
10 F 47 155 53 2000 F
7 G 46 160 50 500 F
8 H 42 163 55 600 F
4 I 18 158 51 200 F
2 J 16 172 52 50 M
1 K 6 120 22 5 F
4)查看和删除重复数据
> duplicated(Familydata$age)
[1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
> duplicated(Familydata$sex)
[1] FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE TRUE
> any(duplicated(Familydata$sex))
[1] TRUE
> table(duplicated(Familydata$sex))
FALSE TRUE
2 9
构造重复值以进行后续操作:使用which()来查看重复值所在行
> Familydata1<-Familydata
> Familydata1[12,]<-Familydata1[2,]
> Familydata1
code age ht wt money sex
1 K 6 120 22 5 F
2 J 16 172 52 50 M
3 A 80 163 71 100 M
4 I 18 158 51 200 F
5 C 69 153 51 300 F
6 B 72 148 60 500 F
7 G 46 160 50 500 F
8 H 42 163 55 600 F
9 D 58 170 67 2000 M
10 F 47 155 53 2000 F
11 E 49 167 64 5000 M
12 J 16 172 52 50 M
> table(duplicated(Familydata1$code))
FALSE TRUE
11 1
> which(duplicated(Familydata1$age)) #查看重复值所在行
[1] 12
删除重复的行:
> #删除重复行
> unique.code<-Familydata1[!duplicated(Familydata1$code),]
> identical(unique.code,Familydata) #比较与原数据是否完全相同
[1] TRUE
5)在数据框中添加和删除变量
添加变量:
> #添加行
> Familydata2<-Familydata
> Familydata2$log10money<-log10(Familydata2$money)
> names(Familydata2)
[1] "code" "age" "ht" "wt" "money" "sex" "log10money"
> Familydata2<-Familydata
> Familydata2<-transform(Familydata2,log10money=log10(money))
> names(Familydata2)
[1] "code" "age" "ht" "wt" "money" "sex" "log10money"
删除变量:
#删除变量
Familydata2[,-7]
Familydata2[-7,]
6)把数据框添加到搜索路径
> #把数据框添加到搜索路径
> attach(Familydata2)
> search()
[1] ".GlobalEnv" "Familydata2" "package:epiDisplay" "package:nnet" "package:MASS"
[6] "package:survival" "package:foreign" "tools:rstudio" "package:stats" "package:graphics"
[11] "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads"
[16] "package:base"
> summary(age) #直接使用变量
Min. 1st Qu. Median Mean 3rd Qu. Max.
6.00 30.00 47.00 45.73 63.50 80.00
> with(infert,summary(age))
Min. 1st Qu. Median Mean 3rd Qu. Max.
21.00 28.00 31.00 31.50 35.25 44.00
> detach(Familydata2) #将不需要的数据框从搜索路径中移除
> search()
[1] ".GlobalEnv" "package:epiDisplay" "package:nnet" "package:MASS" "package:survival"
[6] "package:foreign" "tools:rstudio" "package:stats" "package:graphics" "package:grDevices"
[11] "package:utils" "package:datasets" "package:methods" "Autoloads" "package:base"
二、使用dplyr包处理数据框
dplyr包:以一种统一的规范更高效地处理数据框。
下面以MASS包中的birthwt为例。
#导入birthwt数据
library(dplyr)
data(birthwt,package = "MASS")
birthwt
1)使用slice()和filter()筛选行
slice()操作:按照行号选择行。
> slice(birthwt,2:5)
low age lwt race smoke ptl ht ui ftv bwt
86 0 33 155 3 0 0 0 0 3 2551
87 0 20 105 1 1 0 0 0 1 2557
88 0 21 108 1 1 0 0 1 2 2594
89 0 18 107 1 1 0 0 1 0 2600
filter()筛选操作:
> filter(birthwt,age>35)
low age lwt race smoke ptl ht ui ftv bwt
108 0 36 202 1 0 0 0 0 1 2836
183 0 36 175 1 0 0 0 0 0 3600
226 0 45 123 1 0 0 0 0 1 4990
> filter(birthwt,bwt<2500|bwt>4000)
low age lwt race smoke ptl ht ui ftv bwt
218 0 26 160 3 0 0 0 0 0 4054
219 0 21 115 1 0 0 0 0 1 4054
220 0 22 129 1 0 0 0 0 0 4111
221 0 25 130 1 0 0 0 0 2 4153
222 0 31 120 1 0 0 0 0 2 4167
223 0 35 170 1 0 1 0 0 1 4174
224 0 19 120 1 1 0 0 0 0 4238
.............................................
> filter(birthwt,age>35,bwt<2500|bwt>4000) #逗号分隔多个条件
low age lwt race smoke ptl ht ui ftv bwt
226 0 45 123 1 0 0 0 0 1 4990
2)使用arrange()筛选行
arrange():排序。
arrange(birthwt,bwt) #默认从小到大排序
arrange(birthwt,bwt,age) #当第一个变量相等时,按照第二个变量进行排序
#从大到小排序
arrange(birthwt,desc(bwt))
arrange(birthwt,-bwt)
3)使用select()选择列
> select(birthwt,bwt,age,race,smoke) #后期为避免混淆dplyr::select()
bwt age race smoke
85 2523 19 2 0
86 2551 33 3 0
87 2557 20 1 1
88 2594 21 1 1
89 2600 18 1 1
91 2622 21 3 0
92 2637 22 1 0
4)使用mutate()添加新变量
> mutate(birthwt,lwr.kg=0.4536*lwt) #添加新变量
low age lwt race smoke ptl ht ui ftv bwt lwr.kg
85 0 19 182 2 0 0 0 1 0 2523 82.5552
86 0 33 155 3 0 0 0 0 3 2551 70.3080
87 0 20 105 1 1 0 0 0 1 2557 47.6280
88 0 21 108 1 1 0 0 1 2 2594 48.9888
> mutate(birthwt,lwt=0.4536*lwt) #替换原来的变量
low age lwt race smoke ptl ht ui ftv bwt
85 0 19 82.5552 2 0 0 0 1 0 2523
86 0 33 70.3080 3 0 0 0 0 3 2551
87 0 20 47.6280 1 1 0 0 0 1 2557
88 0 21 48.9888 1 1 0 0 1 2 2594
5)使用summarise()计算统计量
> summarise(birthwt,mwan.bwt=mean(bwt),sd.bwt=sd(bwt))
mwan.bwt sd.bwt
1 2944.587 729.2143
6)使用group_by()拆分数据框
> group_by(birthwt,race)
# A tibble: 189 x 10
# Groups: race [3]
low age lwt race smoke ptl ht ui ftv bwt
<int> <int> <int> <int> <int> <int> <int> <int> <int> <int>
1 0 19 182 2 0 0 0 1 0 2523
2 0 33 155 3 0 0 0 0 3 2551
3 0 20 105 1 1 0 0 0 1 2557
4 0 21 108 1 1 0 0 1 2 2594
5 0 18 107 1 1 0 0 1 0 2600
6 0 21 124 3 0 0 0 0 0 2622
7 0 22 118 1 0 0 0 0 1 2637
8 0 17 103 3 0 0 0 0 1 2637
9 0 29 123 1 1 0 0 0 1 2663
10 0 26 113 1 1 0 0 0 0 2665
# ... with 179 more rows
7)使用传递符‘%>%’组合多个操作
> birthwt %>%
+ mutate(race=factor(race,labels = c("White","Black","Other")))%>%
+ group_by(race)%>%
+ summarise(mean(bwt))
# A tibble: 3 x 2
race `mean(bwt)`
<fct> <dbl>
1 White 3103.
2 Black 2720.
3 Other 2805.
三、数据框的合并
> data1 <- data.frame(id = 1:5,
+ sex = c("female", "male", "male", "female", "male"),
+ age = c(32, 46, 25, 42, 29))
> data1
id sex age
1 1 female 32
2 2 male 46
3 3 male 25
4 4 female 42
5 5 male 29
> data2 <- data.frame(id = 6:10,
+ sex = c("male", "female", "male", "male", "female"),
+ age = c(52, 36, 28, 34, 26))
> data2
id sex age
1 6 male 52
2 7 female 36
3 8 male 28
4 9 male 34
5 10 female 26
1)纵向合并rbind()
> rbind(data1,data2)
id sex age
1 1 female 32
2 2 male 46
3 3 male 25
4 4 female 42
5 5 male 29
6 6 male 52
7 7 female 36
8 8 male 28
9 9 male 34
10 10 female 26
2)横向合并cbind()
> data3 <- data.frame(days = c(28, 57, 15, 7, 19),
+ outcome = c("discharge", "dead", "discharge", "transfer", "discharge"))
> data3
days outcome
1 28 discharge
2 57 dead
3 15 discharge
4 7 transfer
5 19 discharge
> cbind(data1,data3)
id sex age days outcome
1 1 female 32 28 discharge
2 2 male 46 57 dead
3 3 male 25 15 discharge
4 4 female 42 7 transfer
5 5 male 29 19 discharge
3)按照某个共有变量合并
merge():
> data4 <- data.frame(id = c(2, 1, 3, 5, 4),
+ outcome = c("discharge", "dead", "discharge", "transfer", "discharge"))
> data4
id outcome
1 2 discharge
2 1 dead
3 3 discharge
4 5 transfer
5 4 discharge
> mydata<-merge(data1,data4,by="id")
> mydata
id sex age outcome
1 1 female 32 dead
2 2 male 46 discharge
3 3 male 25 discharge
4 4 female 42 discharge
5 5 male 29 transfer
full_join():
> mydata<-full_join(data1,data4,by="id")
> mydata
id sex age outcome
1 1 female 32 dead
2 2 male 46 discharge
3 3 male 25 discharge
4 4 female 42 discharge
5 5 male 29 transfer
四、数据框的长宽格式转换
> data(Indometh)
> wide <- reshape(Indometh, v.names = "conc", idvar = "Subject", timevar = "time", direction = "wide")
> wide
Subject conc.0.25 conc.0.5 conc.0.75 conc.1 conc.1.25 conc.2 conc.3 conc.4 conc.5 conc.6 conc.8
1 1 1.50 0.94 0.78 0.48 0.37 0.19 0.12 0.11 0.08 0.07 0.05
12 2 2.03 1.63 0.71 0.70 0.64 0.36 0.32 0.20 0.25 0.12 0.08
23 3 2.72 1.49 1.16 0.80 0.80 0.39 0.22 0.12 0.11 0.08 0.08
34 4 1.85 1.39 1.02 0.89 0.59 0.40 0.16 0.11 0.10 0.07 0.07
45 5 2.05 1.04 0.81 0.39 0.30 0.23 0.13 0.11 0.08 0.10 0.06
56 6 2.31 1.44 1.03 0.84 0.64 0.42 0.24 0.17 0.13 0.10 0.09
> long <- reshape(wide, idvar = "Subject", varying = list(2:12),
+ v.names = "conc", direction = "long")
> head(long, 12)
Subject time conc
1.1 1 1 1.50
2.1 2 1 2.03
3.1 3 1 2.72
4.1 4 1 1.85
5.1 5 1 2.05
6.1 6 1 2.31
1.2 1 2 0.94
2.2 2 2 1.63
3.2 3 2 1.49
4.2 4 2 1.39
5.2 5 2 1.04
6.2 6 2 1.44
五、缺失值的处理
1)识别缺失值
> height <- c(100, 150, NA, 160)
> height
[1] 100 150 NA 160
> is.na(height)
[1] FALSE FALSE TRUE FALSE
> table(is.na(height))
FALSE TRUE
3 1
任何包含空值的统计量计算结果为NA:
> mean(height)
[1] NA
计算统计量需要移除NA:
> mean(height,na.rm = TRUE)
[1] 136.6667
> mean(na.omit(height))
[1] 136.6667
> summary(height) #summary()会自动忽略NA
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
100.0 125.0 150.0 136.7 155.0 160.0 1
2)探索数据框里的缺失值
missForest包里的函数prodNA():随机生成NA
> library(missForest)
> data(iris)
> set.seed(1234)
> iris.miss<-prodNA(iris)
> summary(iris.miss)
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100 setosa :41
1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300 versicolor:45
Median :5.700 Median :3.000 Median :4.400 Median :1.300 virginica :45
Mean :5.787 Mean :3.059 Mean :3.822 Mean :1.182 NA's :19
3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
NA's :12 NA's :16 NA's :12 NA's :16
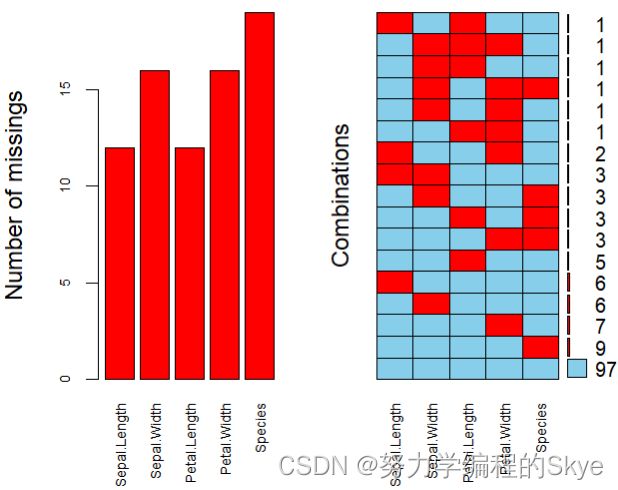
#绘图说明缺失值
library(VIM)
aggr(iris.miss, prop = FALSE, numbers = TRUE, cex.axis = 0.7)
3)填充缺失值
删除缺失值:
> nrow(iris.miss)
[1] 150
> iris.sub<-na.omit(iris.miss)
> nrow(iris.sub)
[1] 97
> iris.sub<-iris.miss[complete.cases(iris.miss),]
> nrow(iris.sub)
[1] 97
使用特定数值替换缺失值:
> iris.miss1<-iris.miss
> Seqal.Length.Mean<-mean(na.omit(iris.miss$Sepal.Length))
> Seqal.Length.Mean
[1] 5.786957
> iris.miss1$Sepal.Length[is.na(iris.miss1$Sepal.Length)]<-Seqal.Length.Mean
> summary((iris$Sepal.Length-iris.miss1$Sepal.Length)/iris$Sepal.Length)
Min. 1st Qu. Median Mean 3rd Qu. Max.
-0.258034 0.000000 0.000000 0.006871 0.000000 0.248447
多重插补:
library(mice)
imputed.data <- mice(iris.miss, seed = 1234)
summary(imputed.data)
imputed.data$imp$Sepal.Length
complete.data <- complete(imputed.data, 3)
summary((iris$Sepal.Length - complete.data$Sepal.Length)/iris$Sepal.Length)
table(iris$Species, complete.data$Species)
六、处理大型数据集的策略
1)清理工作空间
> rm(list=ls(all=TRUE))
PS:其中all默认参数为FALSE,设为TRUE是为了清除包括隐藏对象在内的所有对象。
2)模拟一个大型数据集
> bigdata<-as.data.frame(matrix(rnorm(50000*200),ncol = 200))
> vars<-NULL
> for(i in letters[1:20]){
+ for(j in 1:10){
+ vars<-c(vars,paste(i,j,sep = "_"))
+ }
+ }
> names(bigdata)<-vars
> names(bigdata)
[1] "a_1" "a_2" "a_3" "a_4" "a_5" "a_6" "a_7" "a_8" "a_9" "a_10" "b_1" "b_2" "b_3" "b_4" "b_5"
[16] "b_6" "b_7" "b_8" "b_9" "b_10" "c_1" "c_2" "c_3" "c_4" "c_5" "c_6" "c_7" "c_8" "c_9" "c_10"
[31] "d_1" "d_2" "d_3" "d_4" "d_5" "d_6" "d_7" "d_8" "d_9" "d_10" "e_1" "e_2" "e_3" "e_4" "e_5"
[46] "e_6" "e_7" "e_8" "e_9" "e_10" "f_1" "f_2" "f_3" "f_4" "f_5" "f_6" "f_7" "f_8" "f_9" "f_10"
[61] "g_1" "g_2" "g_3" "g_4" "g_5" "g_6" "g_7" "g_8" "g_9" "g_10" "h_1" "h_2" "h_3" "h_4" "h_5"
[76] "h_6" "h_7" "h_8" "h_9" "h_10" "i_1" "i_2" "i_3" "i_4" "i_5" "i_6" "i_7" "i_8" "i_9" "i_10"
[91] "j_1" "j_2" "j_3" "j_4" "j_5" "j_6" "j_7" "j_8" "j_9" "j_10" "k_1" "k_2" "k_3" "k_4" "k_5"
[106] "k_6" "k_7" "k_8" "k_9" "k_10" "l_1" "l_2" "l_3" "l_4" "l_5" "l_6" "l_7" "l_8" "l_9" "l_10"
[121] "m_1" "m_2" "m_3" "m_4" "m_5" "m_6" "m_7" "m_8" "m_9" "m_10" "n_1" "n_2" "n_3" "n_4" "n_5"
[136] "n_6" "n_7" "n_8" "n_9" "n_10" "o_1" "o_2" "o_3" "o_4" "o_5" "o_6" "o_7" "o_8" "o_9" "o_10"
[151] "p_1" "p_2" "p_3" "p_4" "p_5" "p_6" "p_7" "p_8" "p_9" "p_10" "q_1" "q_2" "q_3" "q_4" "q_5"
[166] "q_6" "q_7" "q_8" "q_9" "q_10" "r_1" "r_2" "r_3" "r_4" "r_5" "r_6" "r_7" "r_8" "r_9" "r_10"
[181] "s_1" "s_2" "s_3" "s_4" "s_5" "s_6" "s_7" "s_8" "s_9" "s_10" "t_1" "t_2" "t_3" "t_4" "t_5"
[196] "t_6" "t_7" "t_8" "t_9" "t_10"
3)剔除不需要的变量
> library(dplyr)
> library(tidyselect)
> subdata1<-select(bigdata,starts_with("a"))
> names(subdata1)
[1] "a_1" "a_2" "a_3" "a_4" "a_5" "a_6" "a_7" "a_8" "a_9" "a_10"
> subdata2<-select(bigdata,ends_with("2"))
> names(subdata2)
[1] "a_2" "b_2" "c_2" "d_2" "e_2" "f_2" "g_2" "h_2" "i_2" "j_2" "k_2" "l_2" "m_2" "n_2" "o_2" "p_2" "q_2" "r_2"
[19] "s_2" "t_2"
> subdata3<-select_at(bigdata,vars(starts_with("a"),starts_with("b")))
> names(subdata3)
[1] "a_1" "a_2" "a_3" "a_4" "a_5" "a_6" "a_7" "a_8" "a_9" "a_10" "b_1" "b_2" "b_3" "b_4" "b_5"
[16] "b_6" "b_7" "b_8" "b_9" "b_10"
> subdata4<-select_at(bigdata,vars(contains("1")))
> names(subdata4)
[1] "a_1" "a_10" "b_1" "b_10" "c_1" "c_10" "d_1" "d_10" "e_1" "e_10" "f_1" "f_10" "g_1" "g_10" "h_1"
[16] "h_10" "i_1" "i_10" "j_1" "j_10" "k_1" "k_10" "l_1" "l_10" "m_1" "m_10" "n_1" "n_10" "o_1" "o_10"
[31] "p_1" "p_10" "q_1" "q_10" "r_1" "r_10" "s_1" "s_10" "t_1" "t_10"
> #剔除1或5结尾的变量
> subdata5<-select_at(bigdata,vars(-contains("1"),-contains("5")))
> names(subdata5)
[1] "a_2" "a_3" "a_4" "a_6" "a_7" "a_8" "a_9" "b_2" "b_3" "b_4" "b_6" "b_7" "b_8" "b_9" "c_2" "c_3" "c_4" "c_6"
[19] "c_7" "c_8" "c_9" "d_2" "d_3" "d_4" "d_6" "d_7" "d_8" "d_9" "e_2" "e_3" "e_4" "e_6" "e_7" "e_8" "e_9" "f_2"
[37] "f_3" "f_4" "f_6" "f_7" "f_8" "f_9" "g_2" "g_3" "g_4" "g_6" "g_7" "g_8" "g_9" "h_2" "h_3" "h_4" "h_6" "h_7"
[55] "h_8" "h_9" "i_2" "i_3" "i_4" "i_6" "i_7" "i_8" "i_9" "j_2" "j_3" "j_4" "j_6" "j_7" "j_8" "j_9" "k_2" "k_3"
[73] "k_4" "k_6" "k_7" "k_8" "k_9" "l_2" "l_3" "l_4" "l_6" "l_7" "l_8" "l_9" "m_2" "m_3" "m_4" "m_6" "m_7" "m_8"
[91] "m_9" "n_2" "n_3" "n_4" "n_6" "n_7" "n_8" "n_9" "o_2" "o_3" "o_4" "o_6" "o_7" "o_8" "o_9" "p_2" "p_3" "p_4"
[109] "p_6" "p_7" "p_8" "p_9" "q_2" "q_3" "q_4" "q_6" "q_7" "q_8" "q_9" "r_2" "r_3" "r_4" "r_6" "r_7" "r_8" "r_9"
[127] "s_2" "s_3" "s_4" "s_6" "s_7" "s_8" "s_9" "t_2" "t_3" "t_4" "t_6" "t_7" "t_8" "t_9"
4)选取数据集的一个随机样本
> sampledata1<-sample_n(subdata5,size=500)
> nrow(sampledata1)
[1] 500
> sampledata2<-sample_frac(subdata5,size=0.02)
> nrow(sampledata2)
[1] 1000