EEG | EEGNet 神经网络分类脑电信号实战(附完整源码)
EEGNet + MNE 分类 Sample数据集
一、环境配置
| Package name | Version |
|---|---|
| Python | 3.7 |
| Tensorflow | 2.7.0 |
| mne | 0.24.1 |
| matplotlib、scikit-learn、numpy | — |
关于 mne ,可以参考我的另一篇博客:MNE-Python | 开源生理信号分析神器(一)
二、数据集介绍
2.1 数据采集
数据集是通过位于MGH/HMS/MIT(麻省总医院)的Athinoula A. Martino 生物医学成像中心的Neuromag Vectorview 系统获得的。同时采集60 通道电极帽的MEG(脑磁图)数据。原始MRI(核磁共振)数据集通过MPRAGE 序列的西门子1.5 T Sonata 扫描仪获取的。数据集下载。
2.2 实验设计
在实验中,受试者的左右视野中会出现棋盘图案,同时会伴随出现在左右耳的音调,刺激间隔为750 ms。此外,在受试者的视野中心会随机出现笑脸图案,受试者被要求在笑脸出现后尽快用右手食指按下按键。实验中刺激和响应的对应关系如图所示。

2.3 数据集目录
Sample 数据集主要包含两个部分:MEG/sample (MEG/EEG 数据)和来自另一位受试者的MRI 重建数据 subjects/sample ,我使用的主要是MEG/sample,其目录如图所示。

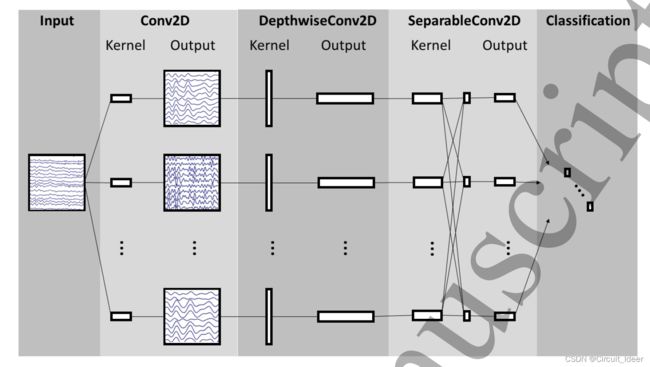
三、网络模型
3.1 网络结构
3.2代码实现
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Dense, Activation, Permute, Dropout
from tensorflow.keras.layers import Conv2D, MaxPooling2D, AveragePooling2D
from tensorflow.keras.layers import SeparableConv2D, DepthwiseConv2D
from tensorflow.keras.layers import BatchNormalization
from tensorflow.keras.layers import SpatialDropout2D
from tensorflow.keras.regularizers import l1_l2
from tensorflow.keras.layers import Input, Flatten
from tensorflow.keras.constraints import max_norm
from tensorflow.keras import backend as K
def EEGNet(nb_classes, Chans = 64, Samples = 128,
dropoutRate = 0.5, kernLength = 64, F1 = 8,
D = 2, F2 = 16, norm_rate = 0.25, dropoutType = 'Dropout'):
if dropoutType == 'SpatialDropout2D':
dropoutType = SpatialDropout2D
elif dropoutType == 'Dropout':
dropoutType = Dropout
else:
raise ValueError('dropoutType must be one of SpatialDropout2D '
'or Dropout, passed as a string.')
input1 = Input(shape = (Chans, Samples, 1))
block1 = Conv2D(F1, (1, kernLength), padding = 'same',
input_shape = (Chans, Samples, 1),
use_bias = False)(input1)
block1 = BatchNormalization()(block1)
block1 = DepthwiseConv2D((Chans, 1), use_bias = False,
depth_multiplier = D,
depthwise_constraint = max_norm(1.))(block1)
block1 = BatchNormalization()(block1)
block1 = Activation('elu')(block1)
block1 = AveragePooling2D((1, 4))(block1)
block1 = dropoutType(dropoutRate)(block1)
block2 = SeparableConv2D(F2, (1, 16),
use_bias = False, padding = 'same')(block1)
block2 = BatchNormalization()(block2)
block2 = Activation('elu')(block2)
block2 = AveragePooling2D((1, 8))(block2)
block2 = dropoutType(dropoutRate)(block2)
flatten = Flatten(name = 'flatten')(block2)
dense = Dense(nb_classes, name = 'dense',
kernel_constraint = max_norm(norm_rate))(flatten)
softmax = Activation('softmax', name = 'softmax')(dense)
return Model(inputs=input1, outputs=softmax)
四、分类实战
4.1 数据集预处理
def get_data4EEGNet():
K.set_image_data_format('channels_last')
# 数据集存放路径(替换成你自己的)
data_path = '/Users/XXX/XXX/'
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
tmin, tmax = -0., 1
event_id = dict(aud_l=1, aud_r=2, vis_l=3, vis_r=4)
kernels, chans, samples = 1, 60, 151
raw = io.Raw(raw_fname, preload=True, verbose=False)
raw.filter(2, None, method='iir')
events = mne.read_events(event_fname)
raw.info['bads'] = ['MEG 2443']
picks = mne.pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=False,
picks=picks, baseline=None, preload=True, verbose=False)
labels = epochs.events[:, -1]
X = epochs.get_data()*1000 # (288,60,151)——(trail数,通道数,采样频率)
y = labels # (288,4)
# 划分训练集、验证集和测试集
X_train = X[0:144,]
Y_train = y[0:144]
X_validate = X[144:216,]
Y_validate = y[144:216]
X_test = X[216:,]
Y_test = y[216:]
Y_train = np_utils.to_categorical(Y_train-1)
Y_validate = np_utils.to_categorical(Y_validate-1)
Y_test = np_utils.to_categorical(Y_test-1)
X_train = X_train.reshape(X_train.shape[0], chans, samples, kernels)
X_validate = X_validate.reshape(X_validate.shape[0], chans, samples, kernels)
X_test = X_test.reshape(X_test.shape[0], chans, samples, kernels)
return X_train, X_validate, X_test, Y_train, Y_validate, Y_test
4.2 完整代码
import numpy as np
import mne
from mne import io
from mne.datasets import sample
from tensorflow.keras import utils as np_utils
from tensorflow.keras.callbacks import ModelCheckpoint
from tensorflow.keras import backend as K
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Dense, Activation, Permute, Dropout
from tensorflow.keras.layers import Conv2D, MaxPooling2D, AveragePooling2D
from tensorflow.keras.layers import SeparableConv2D, DepthwiseConv2D
from tensorflow.keras.layers import BatchNormalization
from tensorflow.keras.layers import SpatialDropout2D
from tensorflow.keras.regularizers import l1_l2
from tensorflow.keras.layers import Input, Flatten
from tensorflow.keras.constraints import max_norm
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
from matplotlib import pyplot as plt
import pathlib
def EEGNet(nb_classes, Chans = 64, Samples = 128,
dropoutRate = 0.5, kernLength = 64, F1 = 8,
D = 2, F2 = 16, norm_rate = 0.25, dropoutType = 'Dropout'):
if dropoutType == 'SpatialDropout2D':
dropoutType = SpatialDropout2D
elif dropoutType == 'Dropout':
dropoutType = Dropout
else:
raise ValueError('dropoutType must be one of SpatialDropout2D '
'or Dropout, passed as a string.')
input1 = Input(shape = (Chans, Samples, 1))
block1 = Conv2D(F1, (1, kernLength), padding = 'same',
input_shape = (Chans, Samples, 1),
use_bias = False)(input1)
block1 = BatchNormalization()(block1)
block1 = DepthwiseConv2D((Chans, 1), use_bias = False,
depth_multiplier = D,
depthwise_constraint = max_norm(1.))(block1)
block1 = BatchNormalization()(block1)
block1 = Activation('elu')(block1)
block1 = AveragePooling2D((1, 4))(block1)
block1 = dropoutType(dropoutRate)(block1)
block2 = SeparableConv2D(F2, (1, 16),
use_bias = False, padding = 'same')(block1)
block2 = BatchNormalization()(block2)
block2 = Activation('elu')(block2)
block2 = AveragePooling2D((1, 8))(block2)
block2 = dropoutType(dropoutRate)(block2)
flatten = Flatten(name = 'flatten')(block2)
dense = Dense(nb_classes, name = 'dense',
kernel_constraint = max_norm(norm_rate))(flatten)
softmax = Activation('softmax', name = 'softmax')(dense)
return Model(inputs=input1, outputs=softmax)
def get_data4EEGNet(kernels, chans, samples):
K.set_image_data_format('channels_last')
data_path = '/Users/XXX'
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
tmin, tmax = -0., 1
event_id = dict(aud_l=1, aud_r=2, vis_l=3, vis_r=4)
raw = io.Raw(raw_fname, preload=True, verbose=False)
raw.filter(2, None, method='iir')
events = mne.read_events(event_fname)
raw.info['bads'] = ['MEG 2443']
picks = mne.pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, proj=False,
picks=picks, baseline=None, preload=True, verbose=False)
labels = epochs.events[:, -1]
X = epochs.get_data()*1000
y = labels
X_train = X[0:144,]
Y_train = y[0:144]
X_validate = X[144:216,]
Y_validate = y[144:216]
X_test = X[216:,]
Y_test = y[216:]
Y_train = np_utils.to_categorical(Y_train-1)
Y_validate = np_utils.to_categorical(Y_validate-1)
Y_test = np_utils.to_categorical(Y_test-1)
X_train = X_train.reshape(X_train.shape[0], chans, samples, kernels)
X_validate = X_validate.reshape(X_validate.shape[0], chans, samples, kernels)
X_test = X_test.reshape(X_test.shape[0], chans, samples, kernels)
return X_train, X_validate, X_test, Y_train, Y_validate, Y_test
kernels, chans, samples = 1, 60, 151
X_train, X_validate, X_test, Y_train, Y_validate, Y_test = get_data4EEGNet(kernels, chans, samples)
model = EEGNet(nb_classes = 4, Chans = chans, Samples = samples,
dropoutRate = 0.5, kernLength = 32, F1 = 8, D = 2, F2 = 16,
dropoutType = 'Dropout')
model.compile(loss='categorical_crossentropy', optimizer='adam',
metrics = ['accuracy'])
checkpointer = ModelCheckpoint(filepath='/Users/XXX/baseline.h5', verbose=1,
save_best_only=True)
class_weights = {0:1, 1:1, 2:1, 3:1}
fittedModel = model.fit(X_train, Y_train, batch_size = 16, epochs = 300,
verbose = 2, validation_data=(X_validate, Y_validate),
callbacks=[checkpointer], class_weight = class_weights)
model.load_weights('./SaveModel/baseline.h5')
probs = model.predict(X_test)
preds = probs.argmax(axis = -1)
acc = np.mean(preds == Y_test.argmax(axis=-1))
print("Classification accuracy: %f " % (acc))
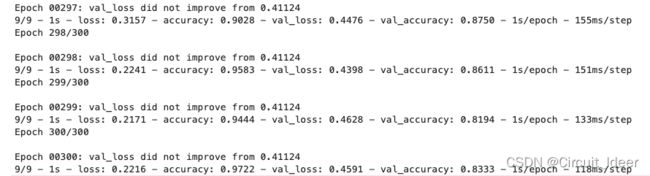
4.3 运行结果
五、参考资料
论文链接: EEGNet: a compact convolutional neural network for EEG-based brain–computer interfaces(Journal of Neural Engineering,SCI JCR2,Impact Factor:4.141)
Github链接: the Army Research Laboratory (ARL) EEGModels project