聚类算法Kmens和密度峰值聚类
目录
Kmeans算法
原理
改进
代码
效果
密度峰值算法
原理
代码
效果
Kmeans算法
原理
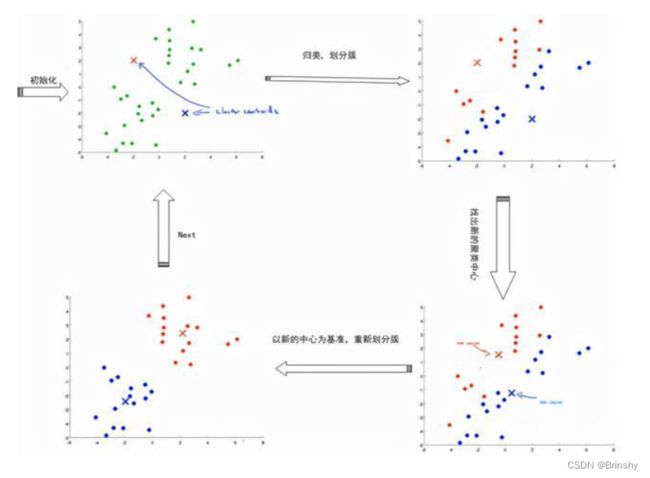
kmeans算法的原理是不断地迭代聚类中心点,主要步骤如下:
(1) 从 n个数据对象任意选择 k 个对象作为初始聚类中心;
(2) 根据每个聚类对象的均值(中心对象),计算每个对象与这些中心对象的距离;并根据最小距离重新对相应对象进行划分;
(3) 重新计算每个(有变化)聚类的均值(中心对象);
(4) 计算标准测度函数,当满足一定条件,如函数收敛时,则算法终止;如果条件不满足则回到步骤(2)。
图片源地址「Python机器学习实战」聚类算法(1)——K-Means聚类|数据点|kmeans_网易订阅
改进
对于数据的处理我们可以利用pca以及熵权法进行一定的降维和权重的改变,使得部分不太合理的数据维数,变得权重更小,部分重要的数据权重变大。从而改善我们的聚类质量。
而利用kmeans++可以使得初始选点更加合理,减少因为初始点位不合引起的聚类暴毙。
其中利用的方法有:
熵权法
PCA降维
利用Kmeans++
代码
from numpy import *
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
import numpy as np
import time
import itertools
INF = 999999999.0
def assess_clustering(list1, list2):
a = b = c = d = 0
for j in range(len(list1)):
if list1[j][0] == list1[j][1] and list2[j][0] == list2[j][1]:
a = a + 1
elif list1[j][0] == list1[j][1] and list2[j][0] != list2[j][1]:
b = b + 1
elif list1[j][0] != list1[j][1] and list2[j][0] == list2[j][1]:
c = c + 1
else:
d = d + 1
jc = a / (a + b + c)
fmi = sqrt(a / (a + b) * a / (a + c))
Rand = (a + d) / len(list1)
return jc, fmi, Rand
def loadDataSet(fileName, splitChar='\t'):
"""
输入:文件名
输出:数据集
描述:从文件读入数据集
"""
dataSet = []
ori_labels = []
with open(fileName) as fr:
for line in fr.readlines():
curline = line.strip().split(splitChar)
ori_labels.append(curline.pop(-1))
fltline = list(map(float, curline))
dataSet.append(fltline)
return dataSet, ori_labels
# 归一化
def Normalization(dataSet, W=[1] * 666):
row = shape(dataSet)[0]
col = shape(dataSet)[1]
sum = 0
mean, si = [], []
for j in range(col):
for i in range(row):
sum += dataSet[i][j]
mean.append(sum / row)
sum = 0
for j in range(col):
for i in range(row):
sum += (dataSet[i][j] - mean[j]) ** 2
si.append(sqrt(sum / row))
sum = 0
for j in range(col):
for i in range(row):
dataSet[i][j] = (dataSet[i][j] - mean[j]) / si[j] * W[j]
return dataSet
def PCA_function(dataSet):
pca = PCA(n_components=2)
pca.fit(np.array(dataSet))
new_dataSet = pca.transform(np.array(dataSet))
k1_spss = pca.components_.T
weight = (np.dot(k1_spss, pca.explained_variance_ratio_)) / np.sum(pca.explained_variance_ratio_)
weighted_weight = weight / np.sum(weight)
return list(new_dataSet), list(weighted_weight)
# 熵权法
def entropy(data0):
# 返回每个样本的指数
# 样本数,指标个数
n, m = np.shape(data0)
# 一行一个样本,一列一个指标
# 下面是归一化
maxium = np.max(data0, axis=0)
minium = np.min(data0, axis=0)
data = (data0 - minium) * 1.0 / (maxium - minium)
##计算第j项指标,第i个样本占该指标的比重
sumzb = np.sum(data, axis=0)
data = data / sumzb
# 对ln0处理
a = data * 1.0
a[np.where(data == 0)] = 0.0001
# #计算每个指标的熵
e = (-1.0 / np.log(n)) * np.sum(data * np.log(a), axis=0)
# #计算权重
w = (1 - e) / np.sum(1 - e)
return w # sha##熵权法
def createDataSet():
"""
输出:数据集
描述:生成数据集
"""
dataSet = [[0, 0], [0, 1], [0, 2], [0, 3], [0, 4],
[0, -1], [0, -2], [0, -3], [0, -4],
[1, 0], [2, 0], [3, 0], [4, 0],
[-1, 0], [-2, 0], [-3, 0], [-4, 0]]
return dataSet
def distEclud(vecA, vecB):
"""
输入:向量A, 向量B
输出:两个向量的欧式距离
"""
return sqrt(sum(power(vecA - vecB, 2)))
def randCent(dataSet, k):
"""
输入:数据集, 聚类个数
输出:k个随机质心的矩阵
"""
n = shape(dataSet)[1]
centroids = mat(zeros((k, n)))
for j in range(n):
minJ = min(dataSet[:, j])
rangeJ = float(max(dataSet[:, j]) - minJ)
centroids[:, j] = minJ + rangeJ * random.rand(k, 1)
return centroids
def nearest(point, cluster_centers):
'''
计算point和cluster_centers之间的最小距离
:param point: 当前的样本点
:param cluster_centers: 当前已经初始化的聚类中心
:return: 返回point与当前聚类中心的最短距离
'''
min_dist = INF
m = np.shape(cluster_centers)[0] # 当前已经初始化聚类中心的个数
for i in range(m):
# 计算point与每个聚类中心之间的距离
d = distEclud(point, cluster_centers[i,])
# 选择最短距离
if min_dist > d:
min_dist = d
return min_dist
def loss(dataSet, clusterAssment, k):
dataSet = mat(dataSet)
num_inside = 0
num_outside = 0
temp = 0
data = []
for i in range(k):
data.append(dataSet[nonzero(clusterAssment[:, 0].A == i)[0], :])
for i in range(len(data)):
for j in range(len(data[i])):
for k in range(len(data[i])):
temp += distEclud(data[i][j], data[i][k]) ** 2
num_inside += temp
temp = 0
for i in range(len(data)):
for j in range(i + 1, len(data)):
for k in range(len(data[i])):
for l in range(len(data[j])):
temp += distEclud(data[i][k], data[j][l])**2
num_outside += temp
temp = 0
return [num_inside, num_outside]
def get_cent(points, k):
'''
kmeans++的初始化聚类中心的方法
:param points: 样本
:param k: 聚类中心的个数
:return: 初始化后的聚类中心
'''
m, n = np.shape(points)
cluster_centers = np.mat(np.zeros((k, n)))
# 1、随机选择一个样本点作为第一个聚类中心
index = np.random.randint(0, m)
cluster_centers[0,] = np.copy(points[index,]) # 复制函数,修改cluster_centers,不会影响points
# 2、初始化一个距离序列
d = [0.0 for _ in range(m)]
for i in range(1, k):
sum_all = 0
for j in range(m):
# 3、对每一个样本找到最近的聚类中心点
d[j] = nearest(points[j,], cluster_centers[0:i, ])
# 4、将所有的最短距离相加
sum_all += d[j]
# 5、取得sum_all之间的随机值
sum_all *= random.rand()
# 6、获得距离最远的样本点作为聚类中心点
for j, di in enumerate(d): # enumerate()函数用于将一个可遍历的数据对象(如列表、元组或字符串)组合为一个索引序列,同事列出数据和数据下标一般用在for循环中
sum_all -= di
if sum_all > 0:
continue
cluster_centers[i] = np.copy(points[j,])
break
return cluster_centers
def kMeans(dataSet, k, distMeans=distEclud, createCent=randCent):
"""
输入:数据集, 聚类个数, 距离计算函数, 生成随机质心函数
输出:质心矩阵, 簇分配和距离矩阵
"""
m = shape(dataSet)[0]
loss_data = [[]]+[[]]
clusterAssment = mat(zeros((m, 2)))
centroids = createCent(dataSet, k)
clusterChanged = True
while clusterChanged:
clusterChanged = False
for i in range(m): # 寻找最近的质心
minDist = INF
minIndex = -1
for j in range(k): # 从k个质心中找出最近的
distJI = distMeans(centroids[j, :], dataSet[i, :])
if distJI < minDist:
minDist = distJI
minIndex = j
if clusterAssment[i, 0] != minIndex:
clusterChanged = True
clusterAssment[i, :] = minIndex, minDist ** 2
for cent in range(k): # 更新第k个质心的位置
ptsInClust = dataSet[nonzero(clusterAssment[:, 0].A == cent)[0]]
centroids[cent, :] = mean(ptsInClust, axis=0)
loss_r = loss(dataSet, clusterAssment, k)
loss_data[0].append(loss_r[0])
loss_data[1].append(loss_r[1])
return centroids, clusterAssment, loss_data
def plotFeature(dataSet, centroids, clusterAssment, loss_data):
m = shape(centroids)[0]
fig = plt.figure()
scatterMarkers = ['s', 'o', '^', '8', 'p', 'd', 'v', 'h', '>', '<']
scatterColors = ['blue', 'green', 'yellow', 'purple', 'orange', 'black', 'brown']
ax = fig.add_subplot(221)
for i in range(m):
ptsInCurCluster = dataSet[nonzero(clusterAssment[:, 0].A == i)[0], :]
markerStyle = scatterMarkers[i % len(scatterMarkers)]
colorSytle = scatterColors[i % len(scatterColors)]
ax.scatter(ptsInCurCluster[:, 0].flatten().A[0], ptsInCurCluster[:, 1].flatten().A[0], marker=markerStyle,
c=colorSytle, s=90)
ax.scatter(centroids[:, 0].flatten().A[0], centroids[:, 1].flatten().A[0], marker='*', c='red', s=100)
# bx = fig.add_subplot(111)
# for i in range(1, 51):
# markerStyle = scatterMarkers[int(clusterAssment.A[i-1][0]) % len(scatterMarkers)]
# colorSytle = scatterColors[int(clusterAssment.A[i-1][0]) % len(scatterColors)]
# bx.scatter(i, 0, marker=markerStyle, c=colorSytle, s=3)
# for i in range(51, 101):
# markerStyle = scatterMarkers[int(clusterAssment.A[i-1][0]) % len(scatterMarkers)]
# colorSytle = scatterColors[int(clusterAssment.A[i-1][0]) % len(scatterColors)]
# bx.scatter(i, 1, marker=markerStyle, c=colorSytle, s=3)
# for i in range(101, 151):
# markerStyle = scatterMarkers[int(clusterAssment.A[i-1][0]) % len(scatterMarkers)]
# colorSytle = scatterColors[int(clusterAssment.A[i-1][0]) % len(scatterColors)]
# bx.scatter(i, 2, marker=markerStyle, c=colorSytle, s=3)
#类内距离和类间距离
cx = fig.add_subplot(222)
x = range(1,len(loss_data[0])+1)
cx.plot(x,loss_data[0])
dx = fig.add_subplot(223)
x = range(1, len(loss_data[1]) + 1)
dx.plot(x, loss_data[1])
def main():
dataSet, ori_labels = loadDataSet('聚类作业/data.txt', splitChar=',')
#dataSet = createDataSet()
W = list(entropy(dataSet)) # W为熵权法权重
# dataSet, W = PCA_function(dataSet)#做pca降维,W为权重,第一个返回参数是降维后的数据集
dataSet = Normalization(dataSet, W) # 归一化
dataSet = mat(dataSet)
best_resultCentroids, best_clustAssing, best_loss_data = None, None, None
best_JC, best_FMI, best_Rand, best_loss = 0, 0, 0, INF
for k in range(1):
resultCentroids, clustAssing, loss_data = kMeans(dataSet, 3, createCent=get_cent) # createCent=get_cent
#计算指标
ans_labels = []
for i in range(len(clustAssing.A)):
ans_labels.append(clustAssing.A[i][0])
comb1 = list(itertools.combinations(ori_labels, 2))
comb2 = list(itertools.combinations(ans_labels, 2))
JC, FMI, Rand = assess_clustering(comb1, comb2)
if best_loss > loss_data[0][-1] / loss_data[1][-1]:
best_resultCentroids, best_clustAssing, best_loss_data = resultCentroids, clustAssing, loss_data
best_loss = loss_data[0][-1] / loss_data[1][-1]
best_JC, best_FMI, best_Rand = JC, FMI, Rand
plotFeature(dataSet, best_resultCentroids, best_clustAssing, best_loss_data)
#聚类的指标参数
print("JC", best_JC)
print("FMI", best_FMI)
print("Rand", best_Rand)
if __name__ == '__main__':
start = time.perf_counter()
main()
end = time.perf_counter()
print('finish all in %s' % str(end - start))
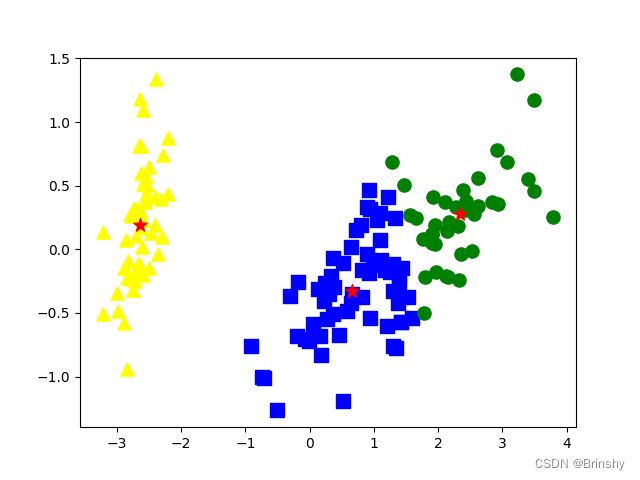
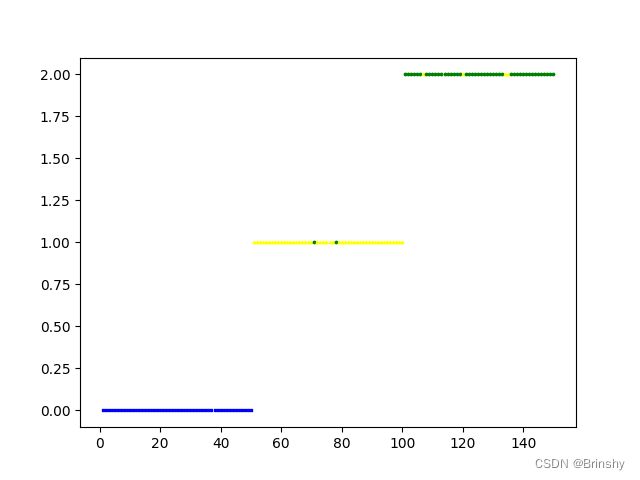
plt.show()效果
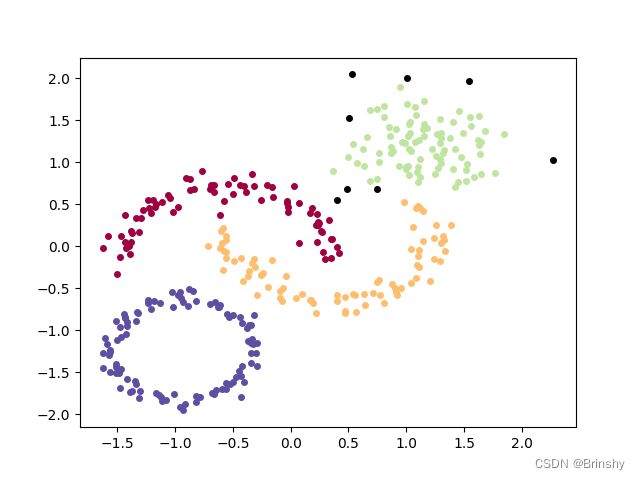
密度峰值算法
原理
第1步,求每个点的密度rho。点的密度就是,以点为中心,以dc为半径,画一个小圆圈,数数里面几个点,圆圈中点的个数就是点的密度。(还可以用高斯核密度求点的密度,求出来的密度是连续型的)
第2步,计算每个点的delta。假设有一个点x,求比点x的密度大的且距离点x最近的那个点y,那么点x与点y之间的距离,就是点x的delta,就这样遍历所有点,把每个点的delta都求出来(注意,delta是距离,谁和谁的距离?x和y的距离,y是谁?y就是比x密度大,且距离x最近的那个点,要满足两个条件,密度比x大且距离最近)
第3步,每个点的密度rho和delta都求出来了,以rho为横坐标,delta为纵坐标,画个二维图,图中右上角的那几个点就是聚类中心,也就是rho和delta都很大的那几个点。(为什么?因为聚类中心有个特点,密度很大,且与密度比它大的点的距离也很大)
第4步,找到聚类中心了,就可以扩展聚类簇了,按照rho从大到小的顺序进行聚类扩展
图片地址为
百度安全验证
代码
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import make_blobs
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import DBSCAN
from sklearn import metrics
import math
import itertools
UNCLASSIFIED = 0
NOISE = -1
# 计算数据点两两之间的距离
def getDistanceMatrix(datas):
N,D = np.shape(datas)
dists = np.zeros([N,N])
for i in range(N):
for j in range(N):
vi = datas[i,:]
vj = datas[j,:]
dists[i,j]= np.sqrt(np.dot((vi-vj),(vi-vj)))
return dists
# 寻找以点cluster_id 为中心,eps 为半径的圆内的所有点的id
def find_points_in_eps(point_id,eps,dists):
index = (dists[point_id]<=eps)
return np.where(index==True)[0].tolist()
# 聚类扩展
# dists : 所有数据两两之间的距离 N x N
# labs : 所有数据的标签 labs N,
# cluster_id : 一个簇的标号
# eps : 密度评估半径
# seeds: 用来进行簇扩展的点
# min_points: 半径内最少的点数
def expand_cluster(dists, labs, cluster_id, seeds, eps, min_points):
i = 0
while i< len(seeds):
# 获取一个临近点
Pn = seeds[i]
# 如果该点被标记为NOISE 则重新标记
if labs[Pn] == NOISE:
labs[Pn] = cluster_id
# 如果该点没有被标记过
elif labs[Pn] == UNCLASSIFIED:
# 进行标记,并计算它的临近点 new_seeds
labs[Pn] = cluster_id
new_seeds = find_points_in_eps(Pn,eps,dists)
# 如果 new_seeds 足够长则把它加入到seed 队列中
if len(new_seeds) >=min_points:
seeds = seeds + new_seeds
i = i+1
def dbscan(datas, eps, min_points):
# 计算 所有点之间的距离
dists = getDistanceMatrix(datas)
# 将所有点的标签初始化为UNCLASSIFIED
n_points = datas.shape[0]
labs = [UNCLASSIFIED]*n_points
cluster_id = 0
# 遍历所有点
for point_id in range(0, n_points):
# 如果当前点已经处理过了
if not(labs[point_id] == UNCLASSIFIED):
continue
# 没有处理过则计算临近点
seeds = find_points_in_eps(point_id,eps,dists)
# 如果临近点数量过少则标记为 NOISE
if len(seeds)