操作记录-2020-12-21: dongfeng_GAS_project
今天老菜鸟接了实验室一个大活,要帮学生把一个课题结掉,因此需要处理三批次数据。当然了,老菜鸟这些天正在忙一些其他事情,不知道这些事情有了结果后,还会不会再继续这些工作了,暂且记录一下自己目前复杂的心情吧。
一、RNA_seq_dongfeng_2020_12_17
1. 建立相应目录
# 新建对应的目录
mkdir raw_data clean_data ballgown bam bam_sort sam fastqc_report GSEA MD5_txt scripts_log
2. 检查数据完整性
这次将提供的md5.txt中数据提前编辑了一下,修改了样品名称和md5中的目录和名称,操作简便多了。
(base) zexing@DNA:~/projects/dongfeng/RNA_seq/2020_12_17/MD5_txt$ cat md5.txt > check_md5sum.txt && md5sum -c check_md5sum.txt
../clean_data/Rescue_WT_2.clean.fq.gz: OK
../clean_data/Rescue_W93A_1.clean.fq.gz: OK
../clean_data/Rescue_WT_1.clean.fq.gz: OK
../clean_data/Rescue_W93A_2.clean.fq.gz: OK
3.在Linux服务器中对RNA_seq数据进行处理
vim新建RNA_seq_script将数据质控、比对、格式转换、排序、拼接和定量综合在一起。
#!/bin/bash
# 上面一行宣告这个script的语法使用bash语法,当程序被执行时,能够载入bash的相关环境配置文件。
# Program
# This program is used for RNA-seq data analysis.
# History
# 2020/12/21 zexing First release
# 设置变量${dir}为常用目录
dir=/f/xudonglab/zexing/projects/dongfeng/RNA_seq/2020_12_17
# 对数据进行质控
fastqc -t 16 -o ${dir}/fastqc_report/ ${dir}/clean_data/*.fq.gz
# 利用for循环进行后续操作
for i in Rescue_W93A Rescue_WT
do
# 对数据进行比对
hisat2 -t -p 16 -x /f/xudonglab/zexing/reference/UCSC_hg19/hisat2_index/hisat2_index_hg19 \
-1 ${dir}/clean_data/${i}_1.clean.fq.gz \
-2 ${dir}/clean_data/${i}_2.clean.fq.gz \
-S ${dir}/sam/${i}.sam
# 对数据进行格式转换
samtools view -@ 16 -S ${dir}/sam/${i}.sam -1b -o ${dir}/bam/${i}.bam
# 对数据进行排序
samtools sort -@ 16 -l 5 -o ${dir}/bam_sort/${i}.bam.sort ${dir}/bam/${i}.bam
# 对数据进行拼接、定量
mkdir ${dir}/ballgown/"$i"
stringtie ${dir}/bam_sort/"$i".bam.sort -o ${dir}/ballgown/"$i"/"$i".gtf \
-p 16 -G /f/xudonglab/zexing/reference/UCSC_hg19/hg19_genes.gtf -e -B \
-A ${dir}/ballgown/"$i"/"$i".gene.tab
done
后台运行RNA_seq_script:
nohup bash RNA_seq_script > RNA_seq_script_log &
4. 使用prepDE.py脚本提取read_counts数值
- 进入ballgown文件夹,将prepDE.py脚本拷贝至当前文件夹
cp /f/xudonglab/zexing/software/prepDE.py ./
- 退出当前conda环境
conda deactivate
- 使用python命令直接运行脚本
python prepDE.py
运行结果中"gene_count_matrix.csv"即是DESeq2的输入文件。
二、RNA_seq_dongfeng_2018_08_16
1. 建立相应目录
# 新建对应的目录
mkdir raw_data clean_data ballgown bam bam_sort sam fastqc_report GSEA MD5_txt scripts_log
2. 在Linux服务器中对RNA_seq数据进行处理
vim新建RNA_seq_script将数据质控、比对、格式转换、排序、拼接和定量综合在一起。
#!/bin/bash
# 上面一行宣告这个script的语法使用bash语法,当程序被执行时,能够载入bash的相关环境配置文件。
# Program
# This program is used for RNA-seq data analysis.
# History
# 2020/12/22 zexing First release
# 设置变量${dir}为常用目录
dir=/f/xudonglab/zexing/projects/dongfeng/RNA_seq/2018_08_16
# 对数据进行质控
fastqc -t 16 -o ${dir}/fastqc_report/ ${dir}/clean_data/*.fq.gz
# 利用for循环进行后续操作
for i in KD_scr_1 KD_sh3_1 KD_sh4_1
do
# 对数据进行比对
hisat2 -t -p 16 -x /f/xudonglab/zexing/reference/UCSC_hg19/hisat2_index/hisat2_index_hg19 \
-U ${dir}/clean_data/${i}.fq.gz \
-S ${dir}/sam/${i}.sam
# 对数据进行格式转换
samtools view -@ 16 -S ${dir}/sam/${i}.sam -1b -o ${dir}/bam/${i}.bam
# 对数据进行排序
samtools sort -@ 16 -l 5 -o ${dir}/bam_sort/${i}.bam.sort ${dir}/bam/${i}.bam
# 对数据进行拼接、定量
mkdir ${dir}/ballgown/"$i"
stringtie ${dir}/bam_sort/"$i".bam.sort -o ${dir}/ballgown/"$i"/"$i".gtf \
-p 16 -G /f/xudonglab/zexing/reference/UCSC_hg19/hg19_genes.gtf -e -B \
-A ${dir}/ballgown/"$i"/"$i".gene.tab
done
后台运行RNA_seq_script:
nohup bash RNA_seq_script > RNA_seq_script_log &
3. 使用prepDE.py脚本提取read_counts数值
- 进入ballgown文件夹,将prepDE.py脚本拷贝至当前文件夹
cp /f/xudonglab/zexing/software/prepDE.py ./
- 退出当前conda环境
conda deactivate
- 使用python命令直接运行脚本
python prepDE.py
运行结果中"gene_count_matrix.csv"即是DESeq2的输入文件。
三、RNA_seq_dongfeng_2021_02_01
1.建立相应目录
# 新建对应的目录
mkdir raw_data clean_data ballgown bam bam_sort sam fastqc_report GSEA MD5_txt scripts_log
2.检查数据完整性
cat md5.txt > check_md5sum.txt && md5sum -c check_md5sum.txt
# 数据处理记录如下
./clean_data/W93A_DOX_1.clean.fq.gz: OK
./clean_data/WT_DOX_2.clean.fq.gz: OK
./clean_data/WT_DOX_1.clean.fq.gz: OK
./clean_data/W93A_DOX_2.clean.fq.gz: OK
md5sum: WARNING: 1 line is improperly formatted
3.在Linux服务器中对RNA_seq数据进行处理
vim新建RNA_seq_script将数据质控、比对、格式转换、排序、拼接和定量综合在一起。
#!/bin/bash
# 上面一行宣告这个script的语法使用bash语法,当程序被执行时,能够载入bash的相关环境配置文件。
# Program
# This program is used for RNA-seq data analysis.
# History
# 2021/02/01 zexing First release
# 设置变量${dir}为常用目录
dir=/f/xudonglab/zexing/projects/dongfeng/RNA_seq/2021_02_01
# 对数据进行质控
fastqc -t 16 -o ${dir}/fastqc_report/ ${dir}/clean_data/*.fq.gz
# 利用for循环进行后续操作
for i in WT_DOX W93A_DOX
do
# 对数据进行比对
hisat2 -t -p 16 -x /f/xudonglab/zexing/reference/UCSC_hg19/hisat2_index/hisat2_index_hg19 \
-1 ${dir}/clean_data/${i}_1.clean.fq.gz \
-2 ${dir}/clean_data/${i}_2.clean.fq.gz \
-S ${dir}/sam/${i}.sam
# 对数据进行格式转换
samtools view -@ 16 -S ${dir}/sam/${i}.sam -1b -o ${dir}/bam/${i}.bam
# 对数据进行排序
samtools sort -@ 16 -l 5 -o ${dir}/bam_sort/${i}.bam.sort ${dir}/bam/${i}.bam
# 对数据进行拼接、定量
mkdir ${dir}/ballgown/"$i"
stringtie ${dir}/bam_sort/"$i".bam.sort -o ${dir}/ballgown/"$i"/"$i".gtf \
-p 16 -G /f/xudonglab/zexing/reference/UCSC_hg19/hg19_genes.gtf -e -B \
-A ${dir}/ballgown/"$i"/"$i".gene.tab
done
后台运行RNA_seq_script:
nohup bash RNA_seq_script > RNA_seq_script_log &
4. 使用prepDE.py脚本提取read_counts数值
- 进入ballgown文件夹,将prepDE.py脚本拷贝至当前文件夹
cp /f/xudonglab/zexing/software/prepDE.py ./
- 退出当前conda环境
conda deactivate
- 使用python命令直接运行脚本
python prepDE.py
运行结果中"gene_count_matrix.csv"即是DESeq2的输入文件。
四、ChIP_seq_dongfeng_2019_10_23
1.建立相应目录
# 新建对应的目录
mkdir raw_data clean_data bam bam_bw bam_sort sam macs2_bdgdiff macs2_callpeak matrix_reference_point matrix_scale_regions fastqc_report MD5_txt scripts_log
2.检查数据完整性
cat md5.txt > check_md5sum.txt && md5sum -c check_md5sum.txt
3. 在Linux服务器中对ChIP_seq数据进行处理并常规call peak。
vim新建ChIP_seq_script_1将数据质控、比对、格式转换、排序、生成目录、bamCoverage命令转换文件格式和常规callpeak综合在一起。
#!/bin/bash
# 上面一行宣告这个script的语法使用bash语法,当程序被执行时,能够载入bash的相关环境配置文件。
# Program
# This program is used for ChIP-seq data analysis.
# History
# 2020/12/22 zexing First release
# 设置变量${dir}为常用目录
dir=/f/xudonglab/zexing/projects/dongfeng/ChIP_seq/2019_10_23 # 用户名称和日期需要更改
input=/f/xudonglab/zexing/reference/ChIP_input/
# 对数据进行质控
fastqc -t 16 -o ${dir}/fastqc_report/ ${dir}/clean_data/*.fq.gz
# 利用for循环进行后续操作
for i in H3K18
do
# 对数据进行比对
bowtie2 -t -p 16 -x /f/xudonglab/zexing/reference/UCSC_hg19/bowtie2_index/hg19 -1 ${dir}/clean_data/${i}_R1.fq.gz -2 ${dir}/clean_data/${i}_R2.fq.gz -S ${dir}/sam/${i}.sam
# 对数据进行格式转换
samtools view -@ 16 -S ${dir}/sam/${i}.sam -1b -o ${dir}/bam/${i}.bam
# 对数据进行排序
samtools sort -@ 16 -l 5 -o ${dir}/bam_sort/${i}.bam.sort ${dir}/bam/${i}.bam
# 对数据生成目录
samtools index -@ 16 ${dir}/bam_sort/${i}.bam.sort
# bamCoverage命令转换文件格式
bamCoverage -p 16 -v -b ${dir}/bam_sort/${i}.bam.sort -o ${dir}/bam_bw/${i}.bam.sort.bw
# 使用macs2进行常规callpeak
macs2 callpeak -t ${dir}/bam_sort/${i}.bam.sort -c ${input}/U87_Normoxia_input.bam.sort -f BAM -g mm -B -q 0.05 --outdir ${dir}/macs2_callpeak/ -n ${i}
done
在后台运行ChIP_seq_script_1:
nohup bash ChIP_seq_script_1 > ChIP_seq_script_1_log &
五、作图分析
1. 对RNA_seq_2018_08_16利用DESeq2包进行差异基因分析
#This script is used for analysis of dongfeng RNA-seq data
#History
# Lizexing 2020-12-23 First release
#genecount文件来源于Stringtie软件分析,后面为本地电脑操作
# DESeq2进行差异分析 ----------------------------------------------------------------
#清空环境变量
rm(list=ls())
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_count/")
#读入基因表达值,设定行名为gene_id
gene_count <- read.csv("gene_count_matrix.csv",stringsAsFactors = F)
#对gene_id一列进行拆分,去除重复名称
library(stringr)
#设置空的"gene_count_1"向量,行数与上面的测序结果一致
gene_count_1<-rep(NA,nrow(gene_count))
#利用for循环,对gene_count数据框中的重复列进行拆分提取
for (i in 1:nrow(gene_count)){
gene_count_1[i] <- unlist(str_split(gene_count[i,1], pattern = "\\|"))[1]
}
#显示拆分后的结果
head(gene_count_1)
#对原数据框中的特定序列重新赋值
gene_count$gene_id <- gene_count_1
#显示文件的前6行信息
head(gene_count)
#将第一列作为文件的行名
rownames(gene_count) <- gene_count[,1]
gene_count <-gene_count[,-1]
#显示文件的前6行信息
head(gene_count)
#将该文件保存至对应目录
write.csv(gene_count, file = "G:/dongfeng/RNA-seq/2018_08_16/gene_count/gene_count.csv", row.names = TRUE)
#加载DESeq2包
library(DESeq2)
#DESeq2需要三种矩阵,分别为countData表达矩阵,colData样品信息矩阵及design差异表达矩阵
#countData为表达矩阵即gene_count
#colData为样品信息矩阵即coldata
#design为差异表达矩阵即批次和条件(对照、处理)等
#设置condition样品组别、重复数
condition <- factor(c(rep("Scr", 1), rep("shGAS41", 2)), levels = c("Scr","shGAS41"))
#显示condition设置信息
condition
#设置group组对应的样品信息矩阵colData
colData <- data.frame(row.names = colnames(gene_count), condition)
#显示colData设置信息
colData
#在R里面用于构建公式对象,~左边为因变量,右边为自变量。
#标准流程:dds <- DESeqDataSetFromMatrix(countData = cts, colData = coldata, design= ~ batch + condition)
#countData为表达矩阵即countdata
#colData为样品信息矩阵即coldata
#design为差异表达矩阵即批次和条件(对照、处理)等
#对dds_group进行标准流程构建
dds <- DESeqDataSetFromMatrix(gene_count, colData, design = ~condition)
#对原始dds_group进行normalize
dds <- DESeq(dds)
#显示dds信息
dds
# 对差异分析结果进行保存 -------------------------------------------------------------
#使用DESeq2包中的results()函数,提取差异分析的结果
#Usage:results(object, contrast, name, .....)
#将提取的差异分析结果定义为变量"res"
#contrast: 定义谁和谁比较,处理组在前,对照组在后
#将group组提取分析结果并保存为res
res = results(dds, contrast=c("condition","shGAS41","Scr"))
#对结果res利用order()函数按pvalue值进行排序
#创建矩阵时,X[i,]指矩阵X中的第i行,X[,j]指矩阵X中的第j列
#order()函数先对数值排序,然后返回排序后各数值的索引,常用用法:V[order(V)]或者df[order(df$variable),]
#对res_group组进行排序
res = res[order(res$pvalue),]
#显示res结果前6行信息
head(res)
#对res_group矩阵进行总结,利用summary命令统计显示一共多少个genes上调和下调
summary(res)
#将差异分析的所有结果进行输出保存
write.csv(res, file="G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/all_different_genes/all_different_genes_genecount.csv")
#利用table函数统计显著差异基因的数目
#显著差异的定义为padj<0.05
table(res$padj<0.05)
#对具有显著性差异的结果进行过滤、提取
#获取padj小于0.05,表达倍数取以2为对数后大于1或者小于-1的差异表达基因(即表达倍数相差2倍及以上)
#使用subset()函数过滤需要的结果至新的变量significant_different_genes_group中
#Usage:subset(x, ...),其中x为objects,...为筛选参数或条件
#对group中数据进行过滤、提取
significant_padj_different_genes <- subset(res, padj < 0.05 & abs(log2FoldChange) > 1)
#使用dim函数查看该结果的维度、规模
dim(significant_padj_different_genes)
#显示结果的前6行信息
head(significant_padj_different_genes)
#对显著差异基因进行输出保存
write.csv(significant_padj_different_genes, file = "G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes/significant_padj_different_genes_genecount.csv")
注意:输出文件保留了pvalue <0.05和padj <0.05两种文件,测序中没有重复,故将两对shRNA的结果作为重复进行分析。
2. 对RNA_seq_2018_08_16绘制火山图
# 火山图 ---------------------------------------------------------------------
#代码参考网站:https://www.jianshu.com/p/e651a182c65d
#火山图的图形非常像火山喷发的形状。
#火山图通常用来展示差异表达的基因,常常出现在芯片、转录组、蛋白组、代谢组等组学检测技术的结果中,并且通常伴随热图一起出现。
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/all_different_genes")
#读取数据至deg.data变量中
#此处需要读取DESeq2分析的全部差异基因,包括显著和非显著基因
#对group_1的全部差异基因进行读取
deg.data <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/all_different_genes/all_different_genes_genecount.csv", header = T, sep = ",")
#更改文件行名称为第一列
rownames(deg.data) <- deg.data$X
#更改文件列名称为需要的名称
colnames(deg.data) <- c("gene_symbol",colnames(deg.data)[c(2:7)])
#显示文件前6行查看文件信息
head(deg.data)
###画火山图只需要其中的log2FC和padj就可以,dongfeng选用pvalue值进行筛选,故此后使用pvalue进行设置
#adj.p.value为校正后的P值,因为基因和基因并不是相互独立的,所以我们需要对P值进行校正来降低结果的假阳性,常用的校正方法为FDR校正
#绘图之前需要对pvalue进行转换,可以拉开差异表达基因之间的间距
#对差异基因pvalue值进行log10转换
deg.data$logP <- -log10(deg.data$pvalue)
#开始绘制基本热图
#利用ggplot2的两个包绘制火山图
#安装ggpubr包、ggthemes包
#install.packages("ggpubr")
#install.packages("ggthemes")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/火山图")
#加载ggpubr包
library(ggpubr)
library(ggthemes)
#x轴为实验组基因表达量比对照组基因表达量的倍数差异
#y轴则为实验组比对照组之后的pvalue值
#火山图上一个点代表一个基因,而颜色则代表他们是显著上调还是显著下调
ggscatter(deg.data, x="log2FoldChange", y="logP") + theme_base()
#上述命令出来的图很丑,需要对log2FoldChange和pvalue数据进行过滤
#新加一列Group
deg.data$Group = "not-significant"
#将pvalue<0.05且log2FC>=1的基因设为显著上调基因
deg.data$Group[which((deg.data$pvalue<0.05) & (deg.data$log2FoldChange >=1))] ="up-regulated"
#将pvalue<0.05且log2FC=<-1的基因设为显著下调基因
deg.data$Group[which((deg.data$pvalue<0.05) & (deg.data$log2FoldChange <= -1))] ="down-regulated"
#查看上调和下调基因数目
table(deg.data$Group)
#使用添加了上调和下调基因的数据重新绘制火山图
#使用color参数指定点的颜色
ggscatter(deg.data, x = "log2FoldChange", y = "logP",
color = "Group") + theme_base()
#修改点的大小size和更改差异表达基因的颜色palette
ggscatter(deg.data, x = "log2FoldChange", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base()
#使用geom_hline和geom_vline分别添加横向和纵向的辅助线
#为火山图添加logP分界线(geom_hline)和logFC分界线(geom_vline)
ggscatter(deg.data, x = "log2FoldChange", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
#为数据增加新的一列Label,将上调和下调差异表达前十的基因绘制在火山图中
#新加一列Label
deg.data$Label = ""
#对差异表达基因的pvalue值进行从小到大排序
deg.data <- deg.data[order(deg.data$pvalue),]
#高表达的基因中,选择pvalue最小的10个
up.genes <- head(deg.data$gene_symbol[which(deg.data$Group == "up-regulated")], 10)
#低表达的基因中,选择pvalue最小的10个
down.genes <- head(deg.data$gene_symbol[which(deg.data$Group == "down-regulated")], 10)
#将up.genes和down.genes合并
deg.top10.genes <- c(as.character(up.genes), as.character(down.genes))
#将top10.gens加入到Label中
deg.data$Label[match(deg.top10.genes, deg.data$gene_symbol)] <- deg.top10.genes
#参数说明:https://www.jianshu.com/p/674f90e020fa
#改变火山图点的颜色和坐标轴标注,使图片更美观
#绘制group1的最终火山图
#对输出的图保存至相应目录
pdf("火山图.pdf")
ggscatter(deg.data, x = "log2FoldChange", y = "logP",
color = "Group",
palette = c("#2f5688", "#BBBBBB", "#CC0000"),
size = 1,
label =deg.data$Label,
font.label = 8,
repel =T,
xlim = c(-15, 15), # x坐标轴的范围
xlab = "log2FoldChange",
ylab = "-log10(P-value)",) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
dev.off()
3. 对RNA_seq_2018_08_16进行GO和KEGG分析
# GO_KEGG -----------------------------------------------------------------
#参考文章:https://www.jianshu.com/p/435d863e0238,
#清空环境变量
rm(list=ls())
#安装包
#BiocManager::install("clusterProfiler")
#BiocManager::install("stringr")
#加载包
library(clusterProfiler)
library(stringr)
library(DOSE)
library(ggplot2)
#clusterProfiler 包里的一些默认作图方法,例如
#barplot(kegg) #富集柱形图
#dotplot(kegg) #富集气泡图
#cnetplot(kegg) #网络图展示富集功能和基因的包含关系
#emapplot(kegg) #网络图展示各富集功能之间共有基因关系
#heatplot(kegg) #热图展示富集功能和基因的包含关系
#Barplot画图参数详解:http://blog.sciencenet.cn/blog-1468811-939797.html
#参考物种的基因注释数据库:人类org.Hs.eg.db,果蝇org.Dm.eg.db,拟南芥org.At.tair.db,小鼠org.Mm.eg.db。
#下载参考人的基因注释库
#BiocManager::install("org.Hs.eg.db")
#加载小鼠的基因注释库
library(org.Hs.eg.db)
#准备输入数据
##待分析的数据就是一串基因名称,可以是ensembl_id、entrze_id或者symbol_id等类型
##读入差异基因的列表(此处根据dongfeng的实验结果,选取significant_gene[pvalue<0.05,abs(log2FC)>=1]进行作图分析
##对于上调、下调基因,需要手动分割成两个单独文件
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes/")
#对样品进行读入
sig.gene_up <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes/significant_pvalue_different_genes_genecount_up.csv")
sig.gene_dn <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes/significant_pvalue_different_genes_genecount_down.csv")
##提取差异基因的列表
gene_up <- sig.gene_up$X
gene_dn <- sig.gene_dn$X
##调整数据格式为字符
gene_up <- as.character(gene_up)
gene_dn <- as.character(gene_dn)
##对基因由SYMBOL转换为ENTREZID格式
##select(x, keys, columns, keytype, ...):基于keys, columns和keytype以data.frame数据类型返回数据,可以是一对多的关系
##mapIds(x, keys, column, keytype, ..., multiVals): 类似于select,只不过就返回一个列。
gene_up.df <- select(org.Hs.eg.db, #人基因注释库
keys = gene_up, #样品组信息
columns = "ENTREZID", #指定基因名称类型
keytype ="SYMBOL") #输入的基因名称类型
gene_dn.df <- select(org.Hs.eg.db, #小鼠基因注释库
keys=gene_dn, #样品组信息
columns = "ENTREZID", #指定基因名称类型
keytype="SYMBOL") #输入的基因名称类型
#GO富集分析
GO_BP_up <- enrichGO(gene= gene_up.df$ENTREZID, #基因列表文件中的基因名称
OrgDb = org.Hs.eg.db, #指定物种的基因数据库,此为人
keyType = "ENTREZID", #指定给定的基因名称类型,此为symbol_id
ont = "BP", #可选BP、MF、CC,也可以指定 ALL 同时计算 3 者
pAdjustMethod = "BH", #指定p值校正方法
pvalueCutoff = 0.05, #指定p值阈值,不显著的值将不显示在结果中
qvalueCutoff = 0.2, #指定q值阈值,不显著的值将不显示在结果中
readable = TRUE) #whether mapping gene ID to gene Name
# minGSSize minimal size of genes annotated by Ontology term for testing.
# maxGSSize maximal size of genes annotated for testing
# pool If ont=’ALL’, whether pool 3 GO sub-ontologies
GO_BP_dn <- enrichGO(gene= gene_dn.df$ENTREZID, #基因列表文件中的基因名称
OrgDb = org.Hs.eg.db, #指定物种的基因数据库,此为小鼠
keyType = "ENTREZID", #指定给定的基因名称类型,此为symbol_id
ont = "BP", #可选BP、MF、CC,也可以指定 ALL 同时计算 3 者
pAdjustMethod = "BH", #指定p值校正方法
pvalueCutoff = 0.05, #指定p值阈值,不显著的值将不显示在结果中
qvalueCutoff = 0.2, #指定q值阈值,不显著的值将不显示在结果中
readable = TRUE) #whether mapping gene ID to gene Name
# minGSSize minimal size of genes annotated by Ontology term for testing.
# maxGSSize maximal size of genes annotated for testing
# pool If ont=’ALL’, whether pool 3 GO sub-ontologies
#对GO富集分析的结果进行输出保存
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/GO_KEGG/")
write.csv(as.data.frame(GO_BP_up), "GO_BP_up.csv")
write.csv(as.data.frame(GO_BP_dn), "GO_BP_dn.csv")
#对GO富集分析进行绘图并输出保存
tiff("GO_BP_up.tiff")
barplot(GO_BP_up, showCategory = 16,title="The GO_BP enrichment analysis of all DEGs ") +
scale_size(range=c(2, 12))+
scale_x_discrete(labels=function(GO_BP_up) str_wrap(GO_BP_up, width = 30))
dev.off()
tiff("GO_BP_dn.tiff")
barplot(GO_BP_dn, showCategory = 16,title="The GO_BP enrichment analysis of all DEGs ") +
scale_size(range=c(2, 12))+
scale_x_discrete(labels=function(GO_BP_dn) str_wrap(GO_BP_dn, width = 30))
dev.off()
#KEGG富集分析
#clusterProfiler的KEGG富集分析方法特殊,它无需加载本地注释库,
#自动使用KEGG的在线数据库进行注释,因此给定的基因名称只能识别entrze id。
#每次打开R计算时,它会自动连接kegg官网获得最近的物种注释信息,因此数据库一定都是最新的
KEGG_up <- enrichKEGG( gene = gene_up.df$ENTREZID, #基因列表文件中的基因名称
keyType = 'kegg', #kegg 富集
organism = 'hsa', #物种,mmu 代表小鼠,hsa代表人类,oas代表绵羊
pAdjustMethod = 'BH', #指定 p 值校正方法
pvalueCutoff = 0.05, #指定 p 值阈值,不显著的值将不显示在结果中
qvalueCutoff = 0.2, #指定 q 值阈值,不显著的值将不显示在结果中
use_internal_data= FALSE )
KEGG_dn <- enrichKEGG( gene = gene_dn.df$ENTREZID, #基因列表文件中的基因名称
keyType = 'kegg', #kegg 富集
organism = 'hsa', #物种,mmu 代表小鼠,hsa代表人类,oas代表绵羊
pAdjustMethod = 'BH', #指定 p 值校正方法
pvalueCutoff = 0.05, #指定 p 值阈值,不显著的值将不显示在结果中
qvalueCutoff = 0.2, #指定 q 值阈值,不显著的值将不显示在结果中
use_internal_data= FALSE )
#KEGG分析结果各列内容:
#ID和Description,富集到的KEGG id和描述;
#GeneRatio和BgRatio,分别为富集到该KEGG条目中的基因数目/给定基因的总数目,以及该条目中背景基因总数目/该物种所有已知的KEGG功能基因数目;
#pvalue、p.adjust和qvalue,p值、校正后p值和q值信息;
#geneID和Count,富集到该KEGG条目中的基因名称(分析中使用的entrze id,故这里也显示的entrze id)和数目。
#如期望显示其它类型的基因id,如通俗的symbol id等类型,由于该分析中只能输入entrze id,因此可以通过基因名称转换的方式对entrze id和symbol id作个匹配转换。
#输出结果
write.table(KEGG_up, 'KEGG_up.csv', sep = ',', quote = FALSE, row.names = FALSE)
write.table(KEGG_dn, 'KEGG_dn.csv', sep = ',', quote = FALSE, row.names = FALSE)
#将输出结果的"ENTREZID"转为"SYMBOL"
#读取数据
#对group_1_up的gene进行名称类型转换
KEGG_up_df <- read.csv("KEGG_up.csv")
#查看文件的整体信息以确认需要转换的列
KEGG_up_df
#with({})函数中的花括号语句,只针对括号内的语句执行,无需担心名字的冲突
#with()函数局限性在于赋值仅在此函数的括号内生效
with(KEGG_up_df, {
gene = NA #对变量gene进行一个定义
for (i in 1:nrow(KEGG_up_df)) #for(i in seq)语句做循环,#nrow()对数据取行数
{
aaa = unlist(str_split(KEGG_up_df$geneID[i],"/")) #str_split函数对字符串进行拆分,unlist函数将拆分的数据合并
gene = c(gene, aaa) #将所有的拆分结果合并在一起
}
KEGG_gene_up <<- gene[-1] #对最开始的gene赋值进行剔除
})
#查看最终提取结果
KEGG_gene_up
#对提取的"ENTREZID"利用select()函数进行转换
#使用birt()函数也可以对ID和symbol进行转换,但是对于重复ID不再进行重复输出
KEGG_gene_up <- select(org.Hs.eg.db, #人基因注释库
keys=KEGG_gene_up, #样品组信息
columns = "SYMBOL", #指定基因名称类型
keytype="ENTREZID") #输入的基因名称类型
#查看最终转换结果
KEGG_gene_up
#对group_1_dn的gene进行名称类型转换
KEGG_dn_df <- read.csv("KEGG_dn.csv")
#查看文件的整体信息以确认需要转换的列
KEGG_dn_df
#with({})函数中的花括号语句,只针对括号内的语句执行,无需担心名字的冲突
#with()函数局限性在于赋值仅在此函数的括号内生效
with(KEGG_dn_df, {
gene = NA #对变量gene进行一个定义
for (i in 1:nrow(KEGG_dn_df)) #for(i in seq)语句做循环,#nrow()对数据取行数
{
aaa = unlist(str_split(KEGG_dn_df$geneID[i],"/")) #str_split函数对字符串进行拆分,unlist函数将拆分的数据合并
gene = c(gene, aaa) #将所有的拆分结果合并在一起
}
KEGG_gene_dn <<- gene[-1] #对最开始的gene赋值进行剔除
})
#查看最终提取结果
KEGG_gene_dn
#对提取的"ENTREZID"利用select()函数进行转换
#使用birt()函数也可以对ID和symbol进行转换,但是对于重复ID不再进行重复输出
KEGG_gene_dn <- select(org.Hs.eg.db, #人基因注释库
keys=KEGG_gene_dn, #样品组信息
columns = "SYMBOL", #指定基因名称类型
keytype="ENTREZID") #输入的基因名称类型
#查看最终转换结果
KEGG_gene_dn
#对转换后的结果输出保存
write.table(KEGG_gene_up, 'KEGG_gene_up.csv', sep = ',', quote = FALSE, row.names = FALSE)
write.table(KEGG_gene_dn, 'KEGG_gene_dn.csv', sep = ',', quote = FALSE, row.names = FALSE)
#对KEGG富集分析的结果进行输出保存
tiff("KEGG_up.tiff")
barplot(KEGG_up, showCategory = 16,title="The KEGG enrichment analysis of all DEGs ") +
scale_size(range=c(2, 12))+
scale_x_discrete(labels=function(KEGG_up) str_wrap(KEGG_up, width = 30))
dev.off()
tiff("KEGG_dn.tiff")
barplot(KEGG_dn, showCategory = 16,title="The KEGG enrichment analysis of all DEGs ") +
scale_size(range=c(2, 12))+
scale_x_discrete(labels=function(KEGG_dn) str_wrap(KEGG_dn, width = 30))
dev.off()
4.对RNA_seq_2018_08_16GO中分析上调的基因对应的promoter区进行motif分析
1. 根据gene_list从hg19_genes.gtf文件中提取gene_TSS信息
也可以使用biomaRt包进行注释,中间出错尚未解决,有机会再尝试。
# 提取待分析的gene_list
# 读入待处理文件
my_file <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes/significant_pvalue_different_genes_genecount_up.csv")
# 根据需要提取gene_symbol
gene_symbol <- my_file$X
# 处理基因组注释文件
# 读入特定基因组文件
hg19_genes_gtf <- read.csv("G:/hg19_genes.gtf", header = FALSE, sep = "\t")
# 对读入的数据某一列进行拆分,此处以空格为例
library(stringr)
# 设置空的"hg19_genes_gtf_V1"向量,其行数与待处理数据行数一致
hg19_genes_gtf_V1<-rep(NA,nrow(hg19_genes_gtf))
hg19_genes_gtf_V2<-rep(NA,nrow(hg19_genes_gtf))
hg19_genes_gtf_V3<-rep(NA,nrow(hg19_genes_gtf))
hg19_genes_gtf_V4<-rep(NA,nrow(hg19_genes_gtf))
# 利用for循环,对hg19_genes_gtf数据框中的第9列进行拆分提取,每一个拆分元素储存至一个变量中
for (i in 1:nrow(hg19_genes_gtf)){
hg19_genes_gtf_V1[i] <- unlist(str_split(hg19_genes_gtf[i,9], pattern = " "))[1]
hg19_genes_gtf_V2[i] <- unlist(str_split(hg19_genes_gtf[i,9], pattern = " "))[2]
hg19_genes_gtf_V3[i] <- unlist(str_split(hg19_genes_gtf[i,9], pattern = " "))[3]
hg19_genes_gtf_V4[i] <- unlist(str_split(hg19_genes_gtf[i,9], pattern = " "))[4]
}
# 对原数据框中的特定序列重新赋值
hg19_genes_gtf <- cbind(hg19_genes_gtf, hg19_genes_gtf_V1, hg19_genes_gtf_V2, hg19_genes_gtf_V3, hg19_genes_gtf_V4)
# 原始gtf文件中字符串末尾有分号故需要删除
# 此处以结尾最后一个字符为例,采用nchar()函数对字符串进行计数后减一,来代表最后一个字符。
# 设置空的"hg19_genes_gtf_V1"向量,其行数与待处理数据行数一致
# 此处NA可以用0代替
hg19_genes_gtf_V5<-rep(NA,nrow(hg19_genes_gtf))
# 利用for循环,对hg19_genes_gtf数据框中的第13列中的分号进行剔除,采用去掉末尾字符的方法
for (i in 1:nrow(hg19_genes_gtf)){
hg19_genes_gtf_V5[i] <- substring(hg19_genes_gtf[i,13], 1, (nchar(as.character(hg19_genes_gtf[i,13]))-1))
}
# 将编辑后的数值合并至原数据框中的特定序列
hg19_genes_gtf <- cbind(hg19_genes_gtf, hg19_genes_gtf_V5)
# 根据目的提取特定列进行保存
hg19_genes_gtf_done <- hg19_genes_gtf[, c(1, 4, 5, 7, 13)]
# 对得到的文件进行输出保存
write.csv(hg19_genes_gtf_done, "G:/hg19_genes_start_end.gtf", quote = FALSE, row.names = FALSE)
##利用match函数提取基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##利用common_gene对各样品进行处理
common_gene_row.NO <- c(match(gene_symbol, hg19_genes_gtf_done$hg19_genes_gtf_V4))
#根据以上行数,对值进行提取
hg19_genes_gtf_pick <- hg19_genes_gtf_done[common_gene_row.NO ,]
# 对于strand -的基因,TSS为较大的数值,对其进行提取
# 设置空的"hg19_genes_gtf_pick_V6"向量,其行数与待处理数据行数一致
hg19_genes_gtf_pick_V6<-rep(0,nrow(hg19_genes_gtf_pick))
hg19_genes_gtf_pick_V7<-rep(0,nrow(hg19_genes_gtf_pick))
# 利用for循环,对hg19_genes_gtf_pick数据框中的两列中的数值进行筛选提取,采用max()/min()函数
for (i in 1:nrow(hg19_genes_gtf_pick)){
if (hg19_genes_gtf_pick[i,4] == "-")
hg19_genes_gtf_pick_V6[i] <- max(hg19_genes_gtf_pick[i,2], hg19_genes_gtf_pick[i,3])
else
hg19_genes_gtf_pick_V7[i] <- min(hg19_genes_gtf_pick[i,2], hg19_genes_gtf_pick[i,3])
}
# 对原数据框中的特定序列重新赋值
hg19_genes_gtf_pick <- cbind(hg19_genes_gtf_pick, hg19_genes_gtf_pick_V6, hg19_genes_gtf_pick_V7)
# 对数据进一步合并处理
hg19_genes_gtf_pick$V8 <- cbind(hg19_genes_gtf_pick$hg19_genes_gtf_pick_V6 + hg19_genes_gtf_pick_V7)
# 对得到的文件进行输出保存
write.csv(hg19_genes_gtf_pick[, c(1, 5, 8)], "G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/GO_KEGG/hg19_genes_start_end.gtf", quote = FALSE, row.names = FALSE)
# 根据自定义对启动子区域进行定义
results <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/GO_KEGG/hg19_genes_start_end.gtf", header = FALSE)
# 定义启动子区域
results$V4 <- cbind(results$V3-3000)
results$V5 <- cbind(results$V3+100)
# 对得到的文件进行输出保存
# 注意此处必须使用tab进行分隔,且不能用冒号,保存后将第一行行名手动删除
write.table(results[, c(1, 4, 5)], "G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/GO_KEGG/results.gff",sep = "\t", quote = FALSE, row.names = FALSE)
2. 根据gene_TSS信息利用bedtools getfast从hg19_genes.fasta文件中提取promoter的序列
参考文章:bedtools----getfasta、最全Bedtools使用说明–只看本文就够了
# 建立promoter_motif文件
# bedtools getfasta 用法介绍
(base) zexing@DNA:~$ bedtools getfasta -h
Tool: bedtools getfasta (aka fastaFromBed)
Version: v2.29.2
Summary: Extract DNA sequences from a fasta file based on feature coordinates.
Usage: bedtools getfasta [OPTIONS] -fi <fasta> -bed <bed/gff/vcf>
Options:
-fi Input FASTA file
-fo Output file (opt., default is STDOUT
-bed BED/GFF/VCF file of ranges to extract from -fi
-name Use the name field and coordinates for the FASTA header
-name+ (deprecated) Use the name field and coordinates for the FASTA header
-nameOnly Use the name field for the FASTA header
-split Given BED12 fmt., extract and concatenate the sequences
from the BED "blocks" (e.g., exons)
-tab Write output in TAB delimited format.
- Default is FASTA format.
-s Force strandedness. If the feature occupies the antisense,
strand, the sequence will be reverse complemented.
- By default, strand information is ignored.
-fullHeader Use full fasta header.
- By default, only the word before the first space or tab
is used.
# 编辑命令如下
bedtools getfasta -fi /f/xudonglab/zexing/reference/UCSC_hg19/hg19.fasta -bed /f/xudonglab/zexing/projects/dongfeng/RNA_seq/2018_08_16/promoter_motif/results.gff -fo /f/xudonglab/zexing/projects/dongfeng/RNA_seq/2018_08_16/promoter_motif/results.fasta
输出结果如下:
 3. 根据gene_promoter信息利用meme软件进行分析
3. 根据gene_promoter信息利用meme软件进行分析
分析软件meme使用conda install命令进行安装,软件使用参考文章:motif分析-MEME
# 命令用法如下
Usage: meme <dataset> [optional arguments]
<dataset> file containing sequences in FASTA format
[-h] print this message
[-o <output dir>] name of directory for output files
will not replace existing directory
[-oc <output dir>] name of directory for output files
will replace existing directory
[-text] output in text format (default is HTML)
[-objfun classic|de|se|cd|ce] objective function (default: classic)
[-test mhg|mbn|mrs] statistical test type (default: mhg)
[-use_llr] use LLR in search for starts in Classic mode
[-neg <negdataset>] file containing control sequences
[-shuf <kmer>] preserve frequencies of k-mers of size <kmer>
when shuffling (default: 2)
[-hsfrac <hsfrac>] fraction of primary sequences in holdout set
(default: 0.5)
[-cefrac <cefrac>] fraction sequence length for CE region
(default: 0.25)
[-searchsize <ssize>] maximum portion of primary dataset to use
for motif search (in characters)
[-maxsize <maxsize>] maximum dataset size in characters
[-norand] do not randomize the order of the input
sequences with -searchsize
[-csites <csites>] maximum number of sites for EM in Classic mode
[-seed <seed>] random seed for shuffling and sampling
[-dna] sequences use DNA alphabet
[-rna] sequences use RNA alphabet
[-protein] sequences use protein alphabet
[-alph <alph file>] sequences use custom alphabet
[-revcomp] allow sites on + or - DNA strands
[-pal] force palindromes (requires -dna)
[-mod oops|zoops|anr] distribution of motifs
[-nmotifs <nmotifs>] maximum number of motifs to find
[-evt <ev>] stop if motif E-value greater than <evt>
[-time <t>] quit before <t> CPU seconds consumed
[-nsites <sites>] number of sites for each motif
[-minsites <minsites>] minimum number of sites for each motif
[-maxsites <maxsites>] maximum number of sites for each motif
[-wnsites <wnsites>] weight on expected number of sites
[-w <w>] motif width
[-minw <minw>] minimum motif width
[-maxw <maxw>] maximum motif width
[-allw] test starts of all widths from minw to maxw
[-nomatrim] do not adjust motif width using multiple
alignment
[-wg <wg>] gap opening cost for multiple alignments
[-ws <ws>] gap extension cost for multiple alignments
[-noendgaps] do not count end gaps in multiple alignments
[-bfile <bfile>] name of background Markov model file
[-markov_order <order>] (maximum) order of Markov model to use or create
[-psp <pspfile>] name of positional priors file
[-maxiter <maxiter>] maximum EM iterations to run
[-distance <distance>] EM convergence criterion
[-prior dirichlet|dmix|mega|megap|addone]
type of prior to use
[-b <b>] strength of the prior
[-plib <plib>] name of Dirichlet prior file
[-spfuzz <spfuzz>] fuzziness of sequence to theta mapping
[-spmap uni|pam] starting point seq to theta mapping type
[-cons <cons>] consensus sequence to start EM from
[-brief <n>] omit sites and sequence tables in
output if more than <n> primary sequences
[-nostatus] do not print progress reports to terminal
[-p <np>] use parallel version with <np> processors
[-sf <sf>] print <sf> as name of sequence file
[-V] verbose mode
[-version] display the version number and exit
# -dna 待测为DNA序列
# -oc 输出路径
# -nostatus 不将软件计算过程输出到屏幕上
# -mod zoops motif的分布类型
# -nmotifs 检测到的motif的最大限制
# -minw motif最小长度
# -maxw motif最大长度
# -objfun classic motif检测的函数算法
# -markov_order 马尔科夫模型使用的顺序
# -oops 每个功能域在每一段序列中都会出现一次,而且只出现一次。这种模式是运算速度最快,而且最为敏感的。但是如果并不是每个序列都包含功能域,那就可能会有不正确的结果。
# -zoops 每个功能域在每一段序列中至多只出现一次,可能不出现。这种模式运算速度较快,敏感性稍弱。
# -anr 每个功能域在每一段序列中出现的次数不定。这种模式运算速度最慢,可能会多花十倍以上的时间。但是对于功能分布的情况完全未知的情况下,这一参数可能会有帮助
# 编辑命令如下
meme /f/xudonglab/zexing/projects/dongfeng/RNA_seq/2018_08_16/promoter_motif/results.fasta -dna -oc /f/xudonglab/zexing/projects/dongfeng/RNA_seq/2018_08_16/promoter_motif/result -nostatus -mod zoops -nmotifs 5 -minw 6 -maxw 20 -objfun classic -markov_order 0
5. 对RNA_seq_2020_12_17利用edgeR包进行差异基因分析
#This script is used for analysis of dongfeng RNA-seq data
#History
# Lizexing 2020-12-23 First release
#genecount文件来源于Stringtie软件分析,后面为本地电脑操作
# edgeR进行无重复转录组差异基因分析 ----------------------------------------------------------------
#参考文章:https://www.jianshu.com/p/4863beb3abf9
#清空环境变量
rm(list=ls())
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_count/")
#读入基因表达值,设定行名为gene_id
gene_count <- read.csv("gene_count_matrix.csv",stringsAsFactors = F)
#对gene_id一列进行拆分,去除重复名称
library(stringr)
#设置空的"gene_count_1"向量,行数与上面的测序结果一致
gene_count_1<-rep(NA,nrow(gene_count))
#利用for循环,对gene_count数据框中的重复列进行拆分提取
for (i in 1:nrow(gene_count)){
gene_count_1[i] <- unlist(str_split(gene_count[i,1], pattern = "\\|"))[1]
}
#显示拆分后的结果
head(gene_count_1)
#对原数据框中的特定序列重新赋值
gene_count$gene_id <- gene_count_1
#显示文件的前6行信息
head(gene_count)
#调整数据顺序
gene_count <-gene_count[, c(1,3,2)]
#显示文件的前6行信息
head(gene_count)
#将该文件保存至对应目录
write.table(gene_count, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_count/gene_count.tab",quote = FALSE, sep = "\t", col.names = TRUE)
#使用edgeR进行无重复差异表达分析
#参考文章:https://www.jianshu.com/p/4863beb3abf9
#BiocManager::install("edgeR")
#启动edgeR
library(edgeR)
#矩阵构建及差异分析
#需要构建2个矩阵:1、表达矩阵;2、分组矩阵( 实验设计);
#表达矩阵
#读取文件
gene_count_2 <- read.csv("G:/dongfeng/RNA-seq/2020_12_17/gene_count/gene_count.tab",sep = "\t", header = TRUE)
data.class(gene_count_2[1,1])
head(gene_count_2)
#构建DGEList对象
#这里因为已经有rawdata的count文件,因此直接用DGEList()函数就行了,否则要用readDGE()函数#安装edgeR
# 构建DGEList
group <- 1:2
y <- DGEList(counts = gene_count_2[,2:3],genes = gene_count_2[,1],group = group)
# 查看构建完y的信息
y
# DGEList对象主要有三部分:
# 1、counts矩阵:包含的是整数counts;
# 2、samples数据框:包含的是文库(sample)信息。包含 lib.size列 :for the library size (sequencing depth) for each sample,如果不自定义, the library sizes will be computed from the column sums of the counts。其中还有一个group列,用于指定每个sample组信息
# 3、一个可选的数据框genes:gene的注释信息
#第二步: 过滤 low counts数据。与DESeq2的预过滤不同,DESeq2的预过滤只是为了改善后续运算性能,在运行过程中依旧会自动处理low count数据,edgeR需要在分析前就要排除那些low count数据,而且非常严格。从生物学角度,有生物学意义的基因的表达量必须高于某一个阈值。从统计学角度上, low count的数据不太可能有显著性差异,而且在多重试验矫正阶段还会拖后腿。
#数据过滤
#由于原来的表达量矩阵基因数太大, 可能存在某些基因根本没有表达, 因此需要预先过滤
keep <- rowSums(cpm(y)>1) >= 1
y <- y[keep, , keep.lib.sizes=FALSE]
y
#这部分代码的意思指的是保留在至少在一个样本里有表达的基因(CPM > 1)。
#标准化
#考虑到测序深度不同, 我们需要对其进行标准化, 避免文库大小不同导致的分析误差.
#edgeR里默认采用TMM(trimmed mean of M-values) 对配对样本进行标准化,用到的函数是calcNormFactors
y <- calcNormFactors(y)
#差异表达分析
#不同差异表达分析工具的目标就是预测出dispersion(离散值), 有了离散值就能够计算p值. 那么dispersion怎么计算呢? edgeR给了几个方法
#根据经验给定一个值(BCV, square-root-dispersion). edgeR给的建议是, 如果你是人类数据, 且实验做的很好(无过多的其他因素影响), 设置为0.4, 如果是遗传上相似的模式物种(这里为小鼠), 设置为0.1 (查询edgeR的bioconductor包所得)
# 如果是小鼠的数据,bcv设定为0.1,如果是人的数据,bcv设定为0.4,如果计算出来的差异基因数量较少,就减小bcv值
y_bcv <- y
bcv <- 0.1
et <- exactTest(y_bcv, dispersion = bcv ^ 2)
gene1 <- decideTestsDGE(et, p.value = 0.05, lfc = 0)
head(gene1)
summary(gene1)
#将结果整理成excel表
# 改一下gene1的名称
colnames(gene1) <- "Signifi"
# 组合将所需要的数据组成一个新的data.frame
results <- cbind(y$genes,y$counts,et$table,gene1)
head(results)
# 将新生成的results数据框写成一个excel数据表
write.csv(x = results,file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/all_different_genes/all_different_genes_genecount.csv",row.names = F)
#对具有显著性差异的结果进行过滤、提取
#获取pvalue小于0.05,表达倍数取以2为对数后大于1或者小于-1的差异表达基因(即表达倍数相差2倍及以上)
#使用subset()函数过滤需要的结果至新的变量significant_different_genes_group中
#Usage:subset(x, ...),其中x为objects,...为筛选参数或条件
#对group中数据进行过滤、提取
significant_pvalue_different_genes <- subset(results, results$PValue < 0.05 & abs(results$logFC) > 1)
#使用dim函数查看该结果的维度、规模
dim(significant_pvalue_different_genes)
#显示结果的前6行信息
head(significant_pvalue_different_genes)
#对显著差异基因进行输出保存
write.csv(significant_pvalue_different_genes, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/significant_different_genes/significant_pvalue_different_genes_genecount.csv")
6. 对RNA_seq_2020_12_17绘制火山图
# 火山图 ---------------------------------------------------------------------
#代码参考网站:https://www.jianshu.com/p/e651a182c65d
#火山图的图形非常像火山喷发的形状。
#火山图通常用来展示差异表达的基因,常常出现在芯片、转录组、蛋白组、代谢组等组学检测技术的结果中,并且通常伴随热图一起出现。
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/all_different_genes")
#读取数据至deg.data变量中
#此处需要读取DESeq2分析的全部差异基因,包括显著和非显著基因
#对全部差异基因进行读取
deg.data <- read.csv("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/all_different_genes/all_different_genes_genecount.csv", header = T, sep = ",")
#更改文件行名称为第一列
rownames(deg.data) <- deg.data$genes
#显示文件前6行查看文件信息
head(deg.data)
###画火山图只需要其中的log2FC和pvalue就可以,dongfeng选用pvalue值进行筛选,故此后使用pvalue进行设置
#adj.p.value为校正后的P值,因为基因和基因并不是相互独立的,所以我们需要对P值进行校正来降低结果的假阳性,常用的校正方法为FDR校正
#绘图之前需要对pvalue进行转换,可以拉开差异表达基因之间的间距
#对差异基因pvalue值进行log10转换
deg.data$logP <- -log10(deg.data$PValue)
#开始绘制基本热图
#利用ggplot2的两个包绘制火山图
#安装ggpubr包、ggthemes包
#install.packages("ggpubr")
#install.packages("ggthemes")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/火山图")
#加载ggpubr包
library(ggpubr)
library(ggthemes)
#x轴为实验组基因表达量比对照组基因表达量的倍数差异
#y轴则为实验组比对照组之后的pvalue值
#火山图上一个点代表一个基因,而颜色则代表他们是显著上调还是显著下调
ggscatter(deg.data, x="logFC", y="logP") + theme_base()
#上述命令出来的图很丑,需要对logFC和pvalue数据进行过滤
#新加一列Group
deg.data$Group = "not-significant"
#将pvalue<0.05且log2FC>=1的基因设为显著上调基因
deg.data$Group[which((deg.data$PValue<0.05) & (deg.data$logFC >=1))] ="up-regulated"
#将pvalue<0.05且log2FC=<-1的基因设为显著下调基因
deg.data$Group[which((deg.data$PValue<0.05) & (deg.data$logFC <= -1))] ="down-regulated"
#查看上调和下调基因数目
table(deg.data$Group)
#使用添加了上调和下调基因的数据重新绘制火山图
#使用color参数指定点的颜色
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group") + theme_base()
#修改点的大小size和更改差异表达基因的颜色palette
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base()
#使用geom_hline和geom_vline分别添加横向和纵向的辅助线
#为火山图添加logP分界线(geom_hline)和logFC分界线(geom_vline)
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
#为数据增加新的一列Label,将上调和下调差异表达前十的基因绘制在火山图中
#新加一列Label
deg.data$Label = ""
#对差异表达基因的pvalue值进行从小到大排序
deg.data <- deg.data[order(deg.data$PValue),]
#高表达的基因中,选择pvalue最小的10个
up.genes <- head(deg.data$genes[which(deg.data$Group == "up-regulated")], 10)
#低表达的基因中,选择pvalue最小的10个
down.genes <- head(deg.data$genes[which(deg.data$Group == "down-regulated")], 10)
#将up.genes和down.genes合并
deg.top10.genes <- c(as.character(up.genes), as.character(down.genes))
#将top10.gens加入到Label中
deg.data$Label[match(deg.top10.genes, deg.data$genes)] <- deg.top10.genes
#参数说明:https://www.jianshu.com/p/674f90e020fa
#改变火山图点的颜色和坐标轴标注,使图片更美观
#绘制group1的最终火山图
#对输出的图保存至相应目录
pdf("火山图.pdf")
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("#2f5688", "#BBBBBB", "#CC0000"),
size = 1,
label =deg.data$Label,
font.label = 8,
repel =T,
xlim = c(-15, 15), # x坐标轴的范围
xlab = "logFC",
ylab = "-log10(P-value)",) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
dev.off()
7. 对RNA_seq_2018_08_16/2020_12_17绘制PCA图
#绘制PCA图
#需要使用各组的FPKM进行绘制
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
#使用intersect函数选取各个样品中的共有gene_id
##intersect函数做地是两个向量的交集,需要先将gene_id转换成字符向量
Scr_gene <- Scr.gene.tab$Gene.ID
shGAS_3_1_gene <- shGAS_3_1.gene.tab$Gene.ID
shGAS_4_1_gene <- shGAS_4_1.gene.tab$Gene.ID
shGAS_WT_1_gene <- shGAS_WT_1.gene.tab$Gene.ID
shGAS_WT_2_gene <- shGAS_WT_2.gene.tab$Gene.ID
shGAS_W93A_1_gene <- shGAS_W93A_1.gene.tab$Gene.ID
shGAS_W93A_2_gene <- shGAS_W93A_2.gene.tab$Gene.ID
##使用intersect函数选取各个样品中的共有gene_id
common_gene_1 <- intersect(Scr_gene, shGAS_3_1_gene)
common_gene_2 <- intersect(common_gene_1, shGAS_4_1_gene)
common_gene_3 <- intersect(common_gene_2, shGAS_WT_1_gene)
common_gene_4 <- intersect(common_gene_3, shGAS_WT_2_gene)
common_gene_5 <- intersect(common_gene_4, shGAS_W93A_1_gene)
common_gene_6 <- intersect(common_gene_5, shGAS_W93A_2_gene)
#利用match函数对共有基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##利用common_gene对各样品进行处理
Scr_common_gene_row.NO <- c(match(common_gene_6, Scr.FPKM$Gene.ID))
shGAS_3_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_common_gene_row.NO <- c(match(common_gene_6, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_common_gene_row.NO <- c(match(common_gene_6, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##利用common_gene对各样品的FPKM值进行提取
Scr_common_gene_FPKM <- Scr.FPKM[Scr_common_gene_row.NO ,]
shGAS_3_1_common_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_common_gene_row.NO ,]
shGAS_4_1_common_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_common_gene_row.NO ,]
shGAS_WT_1_common_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_common_gene_row.NO ,]
shGAS_WT_2_common_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_common_gene_row.NO ,]
shGAS_W93A_1_common_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_common_gene_row.NO ,]
shGAS_W93A_2_common_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_common_gene_row.NO ,]
##利用Merge函数对各样品的FPKM值进行合并
group_common_gene_FPKM <- merge(Scr_common_gene_FPKM, merge(merge(merge(shGAS_3_1_common_gene_FPKM, shGAS_4_1_common_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_common_gene_FPKM, shGAS_WT_2_common_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_common_gene_FPKM, shGAS_W93A_2_common_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
#将gene_id作为文件的行名
rownames(group_common_gene_FPKM) <- group_common_gene_FPKM[,1]
group_common_gene_FPKM <- group_common_gene_FPKM[,-1]
#将FPKM值均为0的基因进行剔除
fpkm <- group_common_gene_FPKM[rowSums(group_common_gene_FPKM>0)>0 , ]
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/PCA/")
#对各样品进行PCA绘图
pca.out=prcomp(t(fpkm), scale=TRUE)
summary(pca.out)
pc12 <- pca.out$x[, 1:2]
pc12 <- data.frame(pc12)
pc12$grp <- c("Scr", "shGAS_3_1", "shGAS_4_1", "shGAS_WT_1", "shGAS_WT_2", "shGAS_W93A_1", "shGAS_W93A_2")
pdf("PCA.pdf")
ggplot(pc12, aes(x=PC1, y=PC2))+geom_point(aes(color=factor(grp)), size=r)+labs(title="PCA visualiztion")
ggplot2::aes(x=PC1, y=PC2)
dev.off()
8. 对RNA_seq_2018_08_16/2020_12_17绘制热图
# heatmap ----------------------------------------------------------------------
#heatmap绘制时,需要使用gene_symbol和FPKM值
#关于FPKM值的说明
#在绘制heatmap热图时,需要使用差异基因所对应的FPKM值,该值的获取有几个办法:
#利用Stringtie的-A参数直接获得
#利用DESeq2的-FPKM参数由count转化获得
#利用ballgown包进行转化获得
#本次采用StringTie的-A参数直接获得
#关于gene_symbol的说明
#对于绘制heatmap图的gene,使用具有差异性的基因进行绘图
#利用之前DESeq2分析得到的abs(log2FoldChange)>=1的基因来做图
#需要根据"significant_different_genes"来从包含FPKM值的文件中将其提取出来,使用到了match()函数
#绘图前的准备工作
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
##将得到的各样本的FPKM值数据保存入heatmap目录
write.table(Scr.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Scr.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_3_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_3_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_4_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_4_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
#对差异基因list进行整理(采用pvalue定义的差异基因进行分析)
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes")
##将各实验组的差异基因导入当前工作环境
group_up <- read.csv("significant_pvalue_different_genes_genecount_up.csv")
group_dn <- read.csv("significant_pvalue_different_genes_genecount_down.csv")
##将各实验组的差异基因list进行提取
group_up_gene <- group_up[,1]
group_dn_gene <- group_dn[,1]
##查看各差异基因名信息
View(group_up_gene)
View(group_dn_gene)
#利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##对group_up各样品进行处理
Scr_group_up_row.NO <- c(match(group_up_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_up_row.NO <- c(match(group_up_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_up_row.NO <- c(match(group_up_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_2.FPKM$Gene.ID))
##对group_dn各样品进行处理
Scr_group_dn_row.NO <- c(match(group_dn_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##对group_up各样品的FPKM值进行提取
Scr_group_up_gene_FPKM <- Scr.FPKM[Scr_group_up_row.NO ,]
shGAS_3_1_group_up_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_up_row.NO ,]
shGAS_4_1_group_up_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_up_row.NO ,]
shGAS_WT_1_group_up_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_up_row.NO ,]
shGAS_WT_2_group_up_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_up_row.NO ,]
shGAS_W93A_1_group_up_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_up_row.NO ,]
shGAS_W93A_2_group_up_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_up_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_up_gene_FPKM)
View(shGAS_3_1_group_up_gene_FPKM)
View(shGAS_4_1_group_up_gene_FPKM)
View(shGAS_WT_1_group_up_gene_FPKM)
View(shGAS_WT_2_group_up_gene_FPKM)
View(shGAS_W93A_1_group_up_gene_FPKM)
View(shGAS_W93A_2_group_up_gene_FPKM)
##对group_dn各样品的FPKM值进行提取
Scr_group_dn_gene_FPKM <- Scr.FPKM[Scr_group_dn_row.NO ,]
shGAS_3_1_group_dn_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_dn_row.NO ,]
shGAS_4_1_group_dn_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_dn_row.NO ,]
shGAS_WT_1_group_dn_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_dn_row.NO ,]
shGAS_WT_2_group_dn_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_dn_row.NO ,]
shGAS_W93A_1_group_dn_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_dn_row.NO ,]
shGAS_W93A_2_group_dn_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_dn_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_dn_gene_FPKM)
View(shGAS_3_1_group_dn_gene_FPKM)
View(shGAS_4_1_group_dn_gene_FPKM)
View(shGAS_WT_1_group_dn_gene_FPKM)
View(shGAS_WT_2_group_dn_gene_FPKM)
View(shGAS_W93A_1_group_dn_gene_FPKM)
View(shGAS_W93A_2_group_dn_gene_FPKM)
#利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
##对group_up各样品的FPKM值进行合并
group_up_gene_FPKM <- merge(Scr_group_up_gene_FPKM, merge(merge(merge(shGAS_3_1_group_up_gene_FPKM, shGAS_4_1_group_up_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_up_gene_FPKM, shGAS_WT_2_group_up_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_up_gene_FPKM, shGAS_W93A_2_group_up_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##对group_dn各样品的FPKM值进行合并
group_dn_gene_FPKM <- merge(Scr_group_dn_gene_FPKM, merge(merge(merge(shGAS_3_1_group_dn_gene_FPKM, shGAS_4_1_group_dn_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_dn_gene_FPKM, shGAS_WT_2_group_dn_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_dn_gene_FPKM, shGAS_W93A_2_group_dn_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##查看合并结果,确认
View(group_up_gene_FPKM)
View(group_dn_gene_FPKM)
#将各实验组差异基因对应的FPKM数据保存至heatmap文件中
write.table(group_up_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_up_gene_FPKM", row.names = FALSE)
write.table(group_dn_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_dn_gene_FPKM", row.names = FALSE)
#开始绘制heatmap图啦啦啦啦啦啦啦啦啦
#代码参考网站:https://www.jianshu.com/p/d86e4afe1065
#安装包(作者说这种方式下载的pheatmap包版本更新一些)
#install.packages('devtools')
#library(devtools)
#install_github("raivokolde/pheatmap")
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/")
#加载包
library(RColorBrewer)#设置颜色用的
library(pheatmap)
#设置配色方案
cc = colorRampPalette(rev(brewer.pal(n=7, name="RdYlBu"))) #Rd=red Yl=yellow Bu=blue
#读入文件,如果确实过多,会无法聚类,最好保证没有缺失,或将缺失替换为0
Group_up<-read.table(file = "group_up_gene_FPKM",row.names = 1,header = T,check.names = F)
Group_dn<-read.table(file = "group_dn_gene_FPKM",row.names = 1,header = T,check.names = F)
#如果矩阵内容是fpkm表达量,一般取log10(fpkm+1)绘图
Group_up=log2(Group_up+1)
Group_dn=log2(Group_dn+1)
#pheatmap参数解释:
#第一个参数是需要用pheatmap画图的数据
#color: 设置颜色。如果想画得更精细一些,可以取cc(1000)
#main: 标题,会显示在最上面
#fontsize: row的字体大小
#scale: 是否归一化为正态分布,可选row,column,none。一般对row进行归一化的情况比较多,column较少。
#border_color: 是否显示边框及边框的颜色,NA不显示, red显示红色。支持简单的颜色单词
#na_col: 设置缺失值的颜色,支持简单颜色单词,一般设置为灰色就满好识别的。
#cluster_rows & cluster_cols: 设置是否对行进行聚类,这个就见仁见智,看你的实际需求了。当缺失值较多的时候是无法进行聚类的。一个解决办法是读取数据的时候不设置缺失值。
#show_rownames & show_colnames: 是否显示行/列的名称
#treeheight_row & treeheight_col: 当前面设置了聚类之后,两边会出现聚类的树,这个参数是设置树的高度的。
#cellheight & cellwidth: 设置每个各自的宽度和高度。有的时候不设置这两个值画出来的树容易放飞自我????
#cutree_row & cutree_col: 是否根据聚类情况把树切开,可以设置切开的份数。
#display_numbers: 设置是否显示每个单元格的值。这个也是个人喜好及文章需求。
#legend: 设置是否显示旁边的bar状图例,emmmm好像还没碰到说不要那个玩意儿的情况。。
#filename: 设置输出文件的名字。可以设置的文件类型有:pdf,png,jpg,tiff,bmp
#绘图group_up
heatmap=pheatmap(Group_up,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_up.tiff")
#绘图group_dn
heatmap=pheatmap(Group_dn,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_dn.tiff")
#采用padj定义的基因进行热图绘制-------------------------------------------------------------------------------------------------------------------------
# heatmap ----------------------------------------------------------------------
#heatmap绘制时,需要使用gene_symbol和FPKM值
#关于FPKM值的说明
#在绘制heatmap热图时,需要使用差异基因所对应的FPKM值,该值的获取有几个办法:
#利用Stringtie的-A参数直接获得
#利用DESeq2的-FPKM参数由count转化获得
#利用ballgown包进行转化获得
#本次采用StringTie的-A参数直接获得
#关于gene_symbol的说明
#对于绘制heatmap图的gene,使用具有差异性的基因进行绘图
#利用之前DESeq2分析得到的abs(log2FoldChange)>=1的基因来做图
#需要根据"significant_different_genes"来从包含FPKM值的文件中将其提取出来,使用到了match()函数
#绘图前的准备工作
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
##将得到的各样本的FPKM值数据保存入heatmap目录
write.table(Scr.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Scr.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_3_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_3_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_4_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_4_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
#对差异基因list进行整理(采用padj定义的差异基因进行分析)
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes")
##将各实验组的差异基因导入当前工作环境
group_up <- read.csv("significant_padj_different_genes_genecount_up.csv")
group_dn <- read.csv("significant_padj_different_genes_genecount_down.csv")
##将各实验组的差异基因list进行提取
group_up_gene <- group_up[,1]
group_dn_gene <- group_dn[,1]
##查看各差异基因名信息
View(group_up_gene)
View(group_dn_gene)
#利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##对group_up各样品进行处理
Scr_group_up_row.NO <- c(match(group_up_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_up_row.NO <- c(match(group_up_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_up_row.NO <- c(match(group_up_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_2.FPKM$Gene.ID))
##对group_dn各样品进行处理
Scr_group_dn_row.NO <- c(match(group_dn_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##对group_up各样品的FPKM值进行提取
Scr_group_up_gene_FPKM <- Scr.FPKM[Scr_group_up_row.NO ,]
shGAS_3_1_group_up_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_up_row.NO ,]
shGAS_4_1_group_up_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_up_row.NO ,]
shGAS_WT_1_group_up_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_up_row.NO ,]
shGAS_WT_2_group_up_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_up_row.NO ,]
shGAS_W93A_1_group_up_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_up_row.NO ,]
shGAS_W93A_2_group_up_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_up_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_up_gene_FPKM)
View(shGAS_3_1_group_up_gene_FPKM)
View(shGAS_4_1_group_up_gene_FPKM)
View(shGAS_WT_1_group_up_gene_FPKM)
View(shGAS_WT_2_group_up_gene_FPKM)
View(shGAS_W93A_1_group_up_gene_FPKM)
View(shGAS_W93A_2_group_up_gene_FPKM)
##对group_dn各样品的FPKM值进行提取
Scr_group_dn_gene_FPKM <- Scr.FPKM[Scr_group_dn_row.NO ,]
shGAS_3_1_group_dn_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_dn_row.NO ,]
shGAS_4_1_group_dn_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_dn_row.NO ,]
shGAS_WT_1_group_dn_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_dn_row.NO ,]
shGAS_WT_2_group_dn_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_dn_row.NO ,]
shGAS_W93A_1_group_dn_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_dn_row.NO ,]
shGAS_W93A_2_group_dn_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_dn_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_dn_gene_FPKM)
View(shGAS_3_1_group_dn_gene_FPKM)
View(shGAS_4_1_group_dn_gene_FPKM)
View(shGAS_WT_1_group_dn_gene_FPKM)
View(shGAS_WT_2_group_dn_gene_FPKM)
View(shGAS_W93A_1_group_dn_gene_FPKM)
View(shGAS_W93A_2_group_dn_gene_FPKM)
#利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
##对group_up各样品的FPKM值进行合并
group_up_gene_FPKM <- merge(Scr_group_up_gene_FPKM, merge(merge(merge(shGAS_3_1_group_up_gene_FPKM, shGAS_4_1_group_up_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_up_gene_FPKM, shGAS_WT_2_group_up_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_up_gene_FPKM, shGAS_W93A_2_group_up_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##对group_dn各样品的FPKM值进行合并
group_dn_gene_FPKM <- merge(Scr_group_dn_gene_FPKM, merge(merge(merge(shGAS_3_1_group_dn_gene_FPKM, shGAS_4_1_group_dn_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_dn_gene_FPKM, shGAS_WT_2_group_dn_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_dn_gene_FPKM, shGAS_W93A_2_group_dn_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##查看合并结果,确认
View(group_up_gene_FPKM)
View(group_dn_gene_FPKM)
#将各实验组差异基因对应的FPKM数据保存至heatmap文件中
write.table(group_up_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_up_gene_padj_FPKM", row.names = FALSE)
write.table(group_dn_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_dn_gene_padj_FPKM", row.names = FALSE)
#开始绘制heatmap图啦啦啦啦啦啦啦啦啦
#代码参考网站:https://www.jianshu.com/p/d86e4afe1065
#安装包(作者说这种方式下载的pheatmap包版本更新一些)
#install.packages('devtools')
#library(devtools)
#install_github("raivokolde/pheatmap")
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/")
#加载包
library(RColorBrewer)#设置颜色用的
library(pheatmap)
#设置配色方案
cc = colorRampPalette(rev(brewer.pal(n=7, name="RdYlBu"))) #Rd=red Yl=yellow Bu=blue
#读入文件,如果确实过多,会无法聚类,最好保证没有缺失,或将缺失替换为0
Group_up<-read.table(file = "group_up_gene_padj_FPKM",row.names = 1,header = T,check.names = F)
Group_dn<-read.table(file = "group_dn_gene_padj_FPKM",row.names = 1,header = T,check.names = F)
#如果矩阵内容是fpkm表达量,一般取log10(fpkm+1)绘图
Group_up=log2(Group_up+1)
Group_dn=log2(Group_dn+1)
#pheatmap参数解释:
#第一个参数是需要用pheatmap画图的数据
#color: 设置颜色。如果想画得更精细一些,可以取cc(1000)
#main: 标题,会显示在最上面
#fontsize: row的字体大小
#scale: 是否归一化为正态分布,可选row,column,none。一般对row进行归一化的情况比较多,column较少。
#border_color: 是否显示边框及边框的颜色,NA不显示, red显示红色。支持简单的颜色单词
#na_col: 设置缺失值的颜色,支持简单颜色单词,一般设置为灰色就满好识别的。
#cluster_rows & cluster_cols: 设置是否对行进行聚类,这个就见仁见智,看你的实际需求了。当缺失值较多的时候是无法进行聚类的。一个解决办法是读取数据的时候不设置缺失值。
#show_rownames & show_colnames: 是否显示行/列的名称
#treeheight_row & treeheight_col: 当前面设置了聚类之后,两边会出现聚类的树,这个参数是设置树的高度的。
#cellheight & cellwidth: 设置每个各自的宽度和高度。有的时候不设置这两个值画出来的树容易放飞自我????
#cutree_row & cutree_col: 是否根据聚类情况把树切开,可以设置切开的份数。
#display_numbers: 设置是否显示每个单元格的值。这个也是个人喜好及文章需求。
#legend: 设置是否显示旁边的bar状图例,emmmm好像还没碰到说不要那个玩意儿的情况。。
#filename: 设置输出文件的名字。可以设置的文件类型有:pdf,png,jpg,tiff,bmp
#绘图group_up
heatmap=pheatmap(Group_up,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_padj_up.tiff")
#绘图group_dn
heatmap=pheatmap(Group_dn,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_padj_dn.tiff")
9. 对ChIP_seq_dongfeng_2019_10_23利用ChIPseeker进行基因注释及转换
此部分使用上一步生成的bed文件在本地机Rstudio中,利用ChIPseeker进行操作。
对常规macs2 callpeak的BED文件进行注释
narrowPeak文件是针对一段peak区域注释,summits.bed是针对peak峰值注释,本次实验中仅仅是查看H3K18cro修饰发生的位置具体对应哪些基因,因此选用narrowPeak文件进行基因注释。
# This script is used for analysis of dongfeng ChIP-seq data
# History
# Lizexing 2020-12-23 First release
# 原始测序数据经过在服务器上进行bowtie2比对和macs2 callpeak分析后,得到的bed文件,
# 将其下载之本地电脑后进行后续操作
# 安装ChIPseeker包
# BiocManager::install("ChIPseeker")
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
# 设置工作目录
setwd("G:/dongfeng/ChIP-seq/2019_10_23/macs2_callpeak/")
#加载ChIPseeker包
library(ChIPseeker)
# 加载基因组注释库
# 安装小鼠注释包
# BiocManager::install("TxDb.Mmusculus.UCSC.mm10.knownGene")
# 安装人的注释包
# BiocManager::install("TxDb.Hsapiens.UCSC.hg19.knownGene")
# 读取chipseq峰的bed文件
H3K18cro_peak <- readPeakFile("G:/dongfeng/ChIP-seq/2019_10_23/macs2_callpeak/H3K18_peaks.narrowPeak")
# 注释,TSS的范围可自定义
# 加载人基因组注释包
require(TxDb.Hsapiens.UCSC.hg19.knownGene)
# 对txdb进行指定
txdb <- TxDb.Hsapiens.UCSC.hg19.knownGene
# 进行注释
H3K18cro_peakAnno <- annotatePeak(H3K18cro_peak, tssRegion = c(-3000, 3000), TxDb = txdb)
# 输出结果
# 设置工作目录
setwd("G:/dongfeng/ChIP-seq/2019_10_23/peakanno/")
write.table(H3K18cro_peakAnno, file = "H3K18cro_peak.csv",sep = '\t', quote = TRUE, row.names = FALSE)
#利用下述脚本将注释文件中的gene_id转化为gene_symbol
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
# 构建待处理基因集的向量
setwd("G:/dongfeng/ChIP-seq/2019_10_23/peakanno/")
H3K18cro_peakAnno_V1 <- read.csv("G:/dongfeng/ChIP-seq/2019_10_23/peakanno/H3K18cro_peak.csv", header = TRUE,sep = "\t" )
# 提取待处理基因集中的gene_id一列并转化为向量格式
H3K18cro_peakAnno_V2 <- as.vector(H3K18cro_peakAnno_V1[, 19])
# BiocManager::install("clusterProfiler")
# 由人的gene_id转化到gene_symbol
library("clusterProfiler")
# BiocManager::install("org.Hs.eg.db")
# BiocManager::install("org.Mm.eg.db")
library("org.Hs.eg.db")
H3K18cro_peakAnno_V3 <- bitr(H3K18cro_peakAnno_V2, # 待转化的文件名
fromType = "ENTREZID", # fromType是指你的数据ID类型是属于哪一类的
toType = "SYMBOL", # toType是指你要转换成哪种ID类型,可以写多种,也可以只写一种
OrgDb = org.Hs.eg.db) # Orgdb是指对应的注释包是哪个
# 查看转化后的结果
View(H3K18cro_peakAnno_V3)
# 保存差异gene_symbol以便后续处理
write.csv(H3K18cro_peakAnno_V3, "G:/dongfeng/ChIP-seq/2019_10_23/peakanno/H3K18cro_peakAnno_V3.txt", quote = FALSE, row.names = FALSE)
10. 绘制GSEA图
- 准备基因表达值FPKM文件
#Gene Set Enrichment Analysis
#参考文章:https://www.jianshu.com/p/aae9b0322b83,
#做差异分析时需要使用全部基因的表达值
#重新提取全部基因的表达值FPKM
#关于FPKM值的说明
#在绘制heatmap热图时,需要使用差异基因所对应的FPKM值,该值的获取有几个办法:
#利用Stringtie的-A参数直接获得
#利用DESeq2的-FPKM参数由count转化获得
#利用ballgown包进行转化获得
#本次采用edgeR的方法获得
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#对RNA_seq_2018_08_16各样品进行处理
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
##将RNA_seq_2018_08_16分析得到的FPKM数据文件导入当前工作环境中
KD_scr_1.tab <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab",sep = "\t", header = TRUE)
KD_sh3_1.tab <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab",sep = "\t", header = TRUE)
KD_sh4_1.tab <- read.csv("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab",sep = "\t", header = TRUE)
##将数据中的FPKM值进行整理
KD_scr_1.FPKM <- KD_scr_1.tab[,c(1,8)]
colnames(KD_scr_1.FPKM)[2] <- "KD_scr_1"
KD_sh3_1.FPKM <- KD_sh3_1.tab[,c(1,8)]
colnames(KD_sh3_1.FPKM)[2] <- "KD_sh3_1"
KD_sh4_1.FPKM <- KD_sh4_1.tab[,c(1,8)]
colnames(KD_sh4_1.FPKM)[2] <- "KD_sh4_1"
##提取各组相同gene_id以便进行合并
common_gene_group1_V1 <- intersect(KD_scr_1.FPKM$Gene.ID, KD_sh3_1.FPKM$Gene.ID)
common_gene_group1_V2 <- intersect(common_gene_group1_V1, KD_sh4_1.FPKM$Gene.ID)
##利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
KD_scr_1_row.NO <- c(match(common_gene_group1_V2, KD_scr_1.FPKM$Gene.ID))
KD_sh3_1_row.NO <- c(match(common_gene_group1_V2, KD_sh3_1.FPKM$Gene.ID))
KD_sh4_1_row.NO <- c(match(common_gene_group1_V2, KD_sh4_1.FPKM$Gene.ID))
##根据以上行数,对各样品的FPKM值进行提取
##对RNA_seq_2018_08_16各样品的FPKM值进行提取
KD_scr_1_gene_FPKM <- na.omit(KD_scr_1.FPKM[KD_scr_1_row.NO ,])
KD_sh3_1_gene_FPKM <- na.omit(KD_sh3_1.FPKM[KD_sh3_1_row.NO ,])
KD_sh4_1_gene_FPKM <- na.omit(KD_sh4_1.FPKM[KD_sh4_1_row.NO ,])
##利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
##对RNA_seq_2018_08_16各样品的FPKM值进行合并
group_1_gene_FPKM <- merge(merge(KD_scr_1_gene_FPKM, KD_sh3_1_gene_FPKM, by="Gene.ID"), KD_sh4_1_gene_FPKM, by="Gene.ID")
##对RNA_seq_2020_12_17各样品进行处理
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
##将2020_12_17分析得到的FPKM数据文件导入当前工作环境中
Rescue_W93A.tab <- read.csv("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A.gene.tab",sep = "\t", header = TRUE)
Rescue_WT.tab <- read.csv("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT.gene.tab",sep = "\t", header = TRUE)
##将数据中的FPKM值进行整理
Rescue_W93A.FPKM <- Rescue_W93A.tab[,c(1,8)]
colnames(Rescue_W93A.FPKM)[2] <- "Rescue_W93A"
Rescue_WT.FPKM <- Rescue_WT.tab[,c(1,8)]
colnames(Rescue_WT.FPKM)[2] <- "Rescue_WT"
##提取各组相同gene_id以便进行合并
common_gene_group2_V1 <- intersect(Rescue_WT.FPKM$Gene.ID, Rescue_W93A.FPKM$Gene.ID)
##利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
Rescue_WT_row.NO <- c(match(common_gene_group2_V1, Rescue_WT.FPKM$Gene.ID))
Rescue_W93A_row.NO <- c(match(common_gene_group2_V1, Rescue_W93A.FPKM$Gene.ID))
##根据以上行数,对各样品的FPKM值进行提取
Rescue_WT_gene_FPKM <- na.omit(Rescue_WT.FPKM[Rescue_WT_row.NO ,])
Rescue_W93A_gene_FPKM <- na.omit(Rescue_W93A.FPKM[Rescue_W93A_row.NO ,])
##利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
group_2_gene_FPKM <- merge(Rescue_WT_gene_FPKM, Rescue_W93A_gene_FPKM, by="Gene.ID")
#将各实验组全部差异基因对应的FPKM数据保存至GSEA文件夹中
##设置工作目录
setwd("G:/dongfeng/ChIP-seq/2019_10_23/GSEA/")
write.table(group_1_gene_FPKM, file = "G:/dongfeng/ChIP-seq/2019_10_23/GSEA/group_1_gene_FPKM.txt", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(group_2_gene_FPKM, file = "G:/dongfeng/ChIP-seq/2019_10_23/GSEA/group_2_gene_FPKM.txt", row.names = FALSE, quote = FALSE, sep = "\t")
- 准备geneset文件即待分析的基因集
#导入特定基因list
##设置工作目录
setwd("G:/dongfeng/ChIP-seq/2019_10_23/peakanno/")
##将各实验组的差异基因导入当前工作环境
geneset <- read.csv("G:/dongfeng/ChIP-seq/2019_10_23/peakanno/H3K18cro_peakAnno_V3.txt")
##将genesymbol进行提取
genesymbol <- geneset[,2]
##查看各差异基因名信息
View(genesymbol)
##对基因名称进行保存
# 保存差异gene_symbol以便后续处理
write.csv(genesymbol, "G:/dongfeng/ChIP-seq/2019_10_23/GSEA/genesymbol.gmx", quote = FALSE, row.names = FALSE)
- 准备分组信息文件
第一行: 样品数目(4)、分组数目(2)、固定数值(1)
第二行: #号必须有、 分组名称
第三行:每个样品对应的分组名称

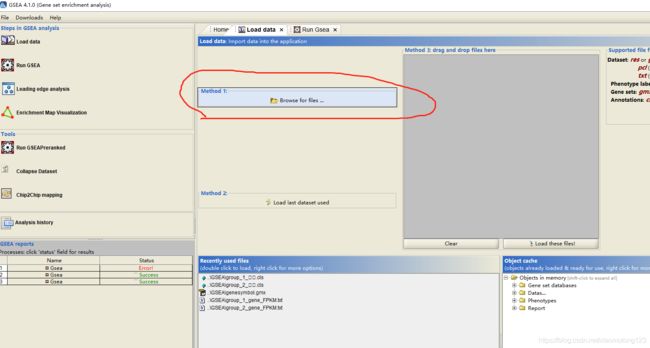
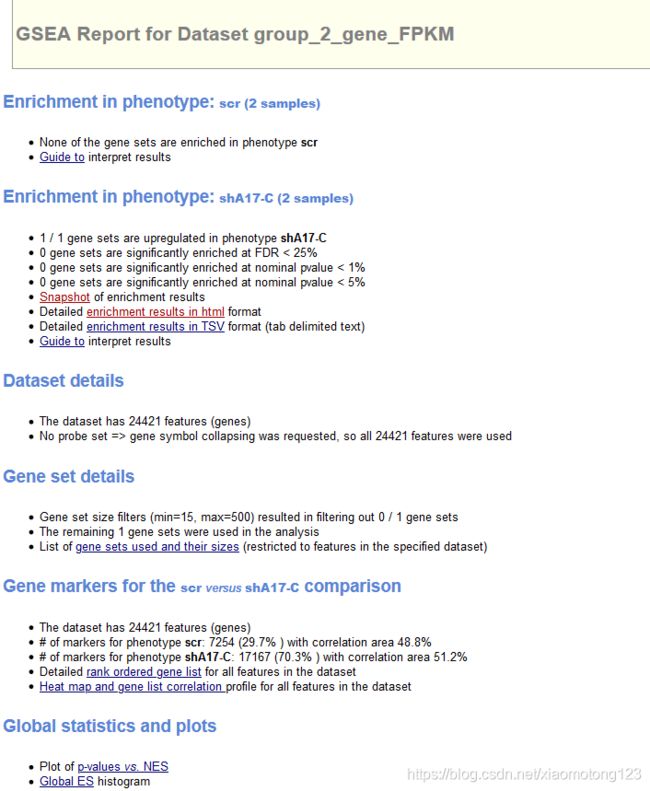
- 利用GSEA软件进行GSEA作图
- 导入基因表达文件(.txt)、分组信息文件(.cls)和geneset文件(.gmx)
11. 对RNA_seq_2021_02_01利用Deseq2进行差异基因分析
#This script is used for analysis of dongfeng RNA-seq data
#History
# Lizexing 2021-02-01 First release
#genecount文件来源于Stringtie软件分析,后面为本地电脑操作
# DESeq2进行差异分析 ----------------------------------------------------------------
#清空环境变量
rm(list=ls())
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2021_02_01/gene_count/")
#读入基因表达值,设定行名为gene_id
gene_count <- read.csv("gene_count_matrix.csv",stringsAsFactors = F)
#对gene_id一列进行拆分,去除重复名称
library(stringr)
#设置空的"gene_count_1"向量,行数与上面的测序结果一致
gene_count_1<-rep(NA,nrow(gene_count))
#利用for循环,对gene_count数据框中的重复列进行拆分提取
for (i in 1:nrow(gene_count)){
gene_count_1[i] <- unlist(str_split(gene_count[i,1], pattern = "\\|"))[1]
}
#显示拆分后的结果
head(gene_count_1)
#对原数据框中的特定序列重新赋值
gene_count$gene_id <- gene_count_1
#显示文件的前6行信息
head(gene_count)
#将第一列作为文件的行名
rownames(gene_count) <- gene_count[,1]
gene_count <-gene_count[,-1]
#显示文件的前6行信息
head(gene_count)
#将该文件保存至对应目录
write.csv(gene_count, file = "G:/dongfeng/RNA-seq/2021_02_01/gene_count/gene_count.csv", row.names = TRUE)
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_count/")
#读入基因表达值,设定行名为gene_id
gene_count_2 <- read.csv("gene_count_matrix.csv",stringsAsFactors = F)
#对gene_id一列进行拆分,去除重复名称
library(stringr)
#设置空的"gene_count_1"向量,行数与上面的测序结果一致
gene_count_1<-rep(NA,nrow(gene_count_2))
#利用for循环,对gene_count数据框中的重复列进行拆分提取
for (i in 1:nrow(gene_count_2)){
gene_count_1[i] <- unlist(str_split(gene_count_2[i,1], pattern = "\\|"))[1]
}
#显示拆分后的结果
head(gene_count_1)
#对原数据框中的特定序列重新赋值
gene_count_2$gene_id <- gene_count_1
#显示文件的前6行信息
head(gene_count_2)
#将第一列作为文件的行名
rownames(gene_count_2) <- gene_count_2[,1]
gene_count_2 <-gene_count_2[,-1]
#显示文件的前6行信息
head(gene_count_2)
#将两次测序结果合并修改名称
gene_count_all <- cbind(gene_count, gene_count_2)
gene_count_all <- gene_count_all[,c(2,4,1,3)]
# BiocManager::install("DESeq2")
#加载DESeq2包
library(DESeq2)
#DESeq2需要三种矩阵,分别为countData表达矩阵,colData样品信息矩阵及design差异表达矩阵
#countData为表达矩阵即gene_count
#colData为样品信息矩阵即coldata
#design为差异表达矩阵即批次和条件(对照、处理)等
#设置condition样品组别、重复数
condition <- factor(c(rep("WT_DOX", 2), rep("W93A_DOX", 2)), levels = c("WT_DOX","W93A_DOX"))
#显示condition设置信息
condition
#设置group组对应的样品信息矩阵colData
colData <- data.frame(row.names = colnames(gene_count_all), condition)
#显示colData设置信息
colData
#在R里面用于构建公式对象,~左边为因变量,右边为自变量。
#标准流程:dds <- DESeqDataSetFromMatrix(countData = cts, colData = coldata, design= ~ batch + condition)
#countData为表达矩阵即countdata
#colData为样品信息矩阵即coldata
#design为差异表达矩阵即批次和条件(对照、处理)等
#对dds_group进行标准流程构建
dds <- DESeqDataSetFromMatrix(gene_count_all, colData, design = ~condition)
#对原始dds_group进行normalize
dds <- DESeq(dds)
#显示dds信息
dds
# 对差异分析结果进行保存 -------------------------------------------------------------
#使用DESeq2包中的results()函数,提取差异分析的结果
#Usage:results(object, contrast, name, .....)
#将提取的差异分析结果定义为变量"res"
#contrast: 定义谁和谁比较,处理组在前,对照组在后
#将group组提取分析结果并保存为res
res = results(dds, contrast=c("condition","W93A_DOX","WT_DOX"))
#对结果res利用order()函数按pvalue值进行排序
#创建矩阵时,X[i,]指矩阵X中的第i行,X[,j]指矩阵X中的第j列
#order()函数先对数值排序,然后返回排序后各数值的索引,常用用法:V[order(V)]或者df[order(df$variable),]
#对res_group组进行排序
res = res[order(res$pvalue),]
#显示res结果前6行信息
head(res)
#对res_group矩阵进行总结,利用summary命令统计显示一共多少个genes上调和下调
summary(res)
#将差异分析的所有结果进行输出保存
write.csv(res, file="G:/dongfeng/RNA-seq/2021_02_01/Rtreatment/all_different_genes/all_different_genes_genecount.csv")
#利用table函数统计显著差异基因的数目
#显著差异的定义为padj<0.05
table(res$pvalue<0.05)
#对具有显著性差异的结果进行过滤、提取
#获取padj小于0.05,表达倍数取以2为对数后大于1或者小于-1的差异表达基因(即表达倍数相差2倍及以上)
#使用subset()函数过滤需要的结果至新的变量significant_different_genes_group中
#Usage:subset(x, ...),其中x为objects,...为筛选参数或条件
#对group中数据进行过滤、提取
significant_pvalue_different_genes <- subset(res, pvalue < 0.05 & abs(log2FoldChange) > 1)
#使用dim函数查看该结果的维度、规模
dim(significant_pvalue_different_genes)
#显示结果的前6行信息
head(significant_pvalue_different_genes)
#对显著差异基因进行输出保存
write.csv(significant_pvalue_different_genes, file = "G:/dongfeng/RNA-seq/2021_02_01/Rtreatment/significant_different_genes/significant_pvalue_different_genes_genecount.csv")
6. 对RNA_seq_2020_12_17绘制火山图
# 火山图 ---------------------------------------------------------------------
#代码参考网站:https://www.jianshu.com/p/e651a182c65d
#火山图的图形非常像火山喷发的形状。
#火山图通常用来展示差异表达的基因,常常出现在芯片、转录组、蛋白组、代谢组等组学检测技术的结果中,并且通常伴随热图一起出现。
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/all_different_genes")
#读取数据至deg.data变量中
#此处需要读取DESeq2分析的全部差异基因,包括显著和非显著基因
#对全部差异基因进行读取
deg.data <- read.csv("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/all_different_genes/all_different_genes_genecount.csv", header = T, sep = ",")
#更改文件行名称为第一列
rownames(deg.data) <- deg.data$genes
#显示文件前6行查看文件信息
head(deg.data)
###画火山图只需要其中的log2FC和pvalue就可以,dongfeng选用pvalue值进行筛选,故此后使用pvalue进行设置
#adj.p.value为校正后的P值,因为基因和基因并不是相互独立的,所以我们需要对P值进行校正来降低结果的假阳性,常用的校正方法为FDR校正
#绘图之前需要对pvalue进行转换,可以拉开差异表达基因之间的间距
#对差异基因pvalue值进行log10转换
deg.data$logP <- -log10(deg.data$PValue)
#开始绘制基本热图
#利用ggplot2的两个包绘制火山图
#安装ggpubr包、ggthemes包
#install.packages("ggpubr")
#install.packages("ggthemes")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/火山图")
#加载ggpubr包
library(ggpubr)
library(ggthemes)
#x轴为实验组基因表达量比对照组基因表达量的倍数差异
#y轴则为实验组比对照组之后的pvalue值
#火山图上一个点代表一个基因,而颜色则代表他们是显著上调还是显著下调
ggscatter(deg.data, x="logFC", y="logP") + theme_base()
#上述命令出来的图很丑,需要对logFC和pvalue数据进行过滤
#新加一列Group
deg.data$Group = "not-significant"
#将pvalue<0.05且log2FC>=1的基因设为显著上调基因
deg.data$Group[which((deg.data$PValue<0.05) & (deg.data$logFC >=1))] ="up-regulated"
#将pvalue<0.05且log2FC=<-1的基因设为显著下调基因
deg.data$Group[which((deg.data$PValue<0.05) & (deg.data$logFC <= -1))] ="down-regulated"
#查看上调和下调基因数目
table(deg.data$Group)
#使用添加了上调和下调基因的数据重新绘制火山图
#使用color参数指定点的颜色
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group") + theme_base()
#修改点的大小size和更改差异表达基因的颜色palette
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base()
#使用geom_hline和geom_vline分别添加横向和纵向的辅助线
#为火山图添加logP分界线(geom_hline)和logFC分界线(geom_vline)
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("green", "gray", "red"),
size = 1) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
#为数据增加新的一列Label,将上调和下调差异表达前十的基因绘制在火山图中
#新加一列Label
deg.data$Label = ""
#对差异表达基因的pvalue值进行从小到大排序
deg.data <- deg.data[order(deg.data$PValue),]
#高表达的基因中,选择pvalue最小的10个
up.genes <- head(deg.data$genes[which(deg.data$Group == "up-regulated")], 10)
#低表达的基因中,选择pvalue最小的10个
down.genes <- head(deg.data$genes[which(deg.data$Group == "down-regulated")], 10)
#将up.genes和down.genes合并
deg.top10.genes <- c(as.character(up.genes), as.character(down.genes))
#将top10.gens加入到Label中
deg.data$Label[match(deg.top10.genes, deg.data$genes)] <- deg.top10.genes
#参数说明:https://www.jianshu.com/p/674f90e020fa
#改变火山图点的颜色和坐标轴标注,使图片更美观
#绘制group1的最终火山图
#对输出的图保存至相应目录
pdf("火山图.pdf")
ggscatter(deg.data, x = "logFC", y = "logP",
color = "Group",
palette = c("#2f5688", "#BBBBBB", "#CC0000"),
size = 1,
label =deg.data$Label,
font.label = 8,
repel =T,
xlim = c(-15, 15), # x坐标轴的范围
xlab = "logFC",
ylab = "-log10(P-value)",) + theme_base() +
geom_hline(yintercept = 1.30, linetype="dashed") +
geom_vline(xintercept = c(-1,1), linetype="dashed")
dev.off()
7. 对RNA_seq_2018_08_16/2020_12_17绘制PCA图
#绘制PCA图
#需要使用各组的FPKM进行绘制
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
#使用intersect函数选取各个样品中的共有gene_id
##intersect函数做地是两个向量的交集,需要先将gene_id转换成字符向量
Scr_gene <- Scr.gene.tab$Gene.ID
shGAS_3_1_gene <- shGAS_3_1.gene.tab$Gene.ID
shGAS_4_1_gene <- shGAS_4_1.gene.tab$Gene.ID
shGAS_WT_1_gene <- shGAS_WT_1.gene.tab$Gene.ID
shGAS_WT_2_gene <- shGAS_WT_2.gene.tab$Gene.ID
shGAS_W93A_1_gene <- shGAS_W93A_1.gene.tab$Gene.ID
shGAS_W93A_2_gene <- shGAS_W93A_2.gene.tab$Gene.ID
##使用intersect函数选取各个样品中的共有gene_id
common_gene_1 <- intersect(Scr_gene, shGAS_3_1_gene)
common_gene_2 <- intersect(common_gene_1, shGAS_4_1_gene)
common_gene_3 <- intersect(common_gene_2, shGAS_WT_1_gene)
common_gene_4 <- intersect(common_gene_3, shGAS_WT_2_gene)
common_gene_5 <- intersect(common_gene_4, shGAS_W93A_1_gene)
common_gene_6 <- intersect(common_gene_5, shGAS_W93A_2_gene)
#利用match函数对共有基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##利用common_gene对各样品进行处理
Scr_common_gene_row.NO <- c(match(common_gene_6, Scr.FPKM$Gene.ID))
shGAS_3_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_common_gene_row.NO <- c(match(common_gene_6, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_common_gene_row.NO <- c(match(common_gene_6, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_common_gene_row.NO <- c(match(common_gene_6, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##利用common_gene对各样品的FPKM值进行提取
Scr_common_gene_FPKM <- Scr.FPKM[Scr_common_gene_row.NO ,]
shGAS_3_1_common_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_common_gene_row.NO ,]
shGAS_4_1_common_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_common_gene_row.NO ,]
shGAS_WT_1_common_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_common_gene_row.NO ,]
shGAS_WT_2_common_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_common_gene_row.NO ,]
shGAS_W93A_1_common_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_common_gene_row.NO ,]
shGAS_W93A_2_common_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_common_gene_row.NO ,]
##利用Merge函数对各样品的FPKM值进行合并
group_common_gene_FPKM <- merge(Scr_common_gene_FPKM, merge(merge(merge(shGAS_3_1_common_gene_FPKM, shGAS_4_1_common_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_common_gene_FPKM, shGAS_WT_2_common_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_common_gene_FPKM, shGAS_W93A_2_common_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
#将gene_id作为文件的行名
rownames(group_common_gene_FPKM) <- group_common_gene_FPKM[,1]
group_common_gene_FPKM <- group_common_gene_FPKM[,-1]
#将FPKM值均为0的基因进行剔除
fpkm <- group_common_gene_FPKM[rowSums(group_common_gene_FPKM>0)>0 , ]
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/PCA/")
#对各样品进行PCA绘图
pca.out=prcomp(t(fpkm), scale=TRUE)
summary(pca.out)
pc12 <- pca.out$x[, 1:2]
pc12 <- data.frame(pc12)
pc12$grp <- c("Scr", "shGAS_3_1", "shGAS_4_1", "shGAS_WT_1", "shGAS_WT_2", "shGAS_W93A_1", "shGAS_W93A_2")
pdf("PCA.pdf")
ggplot(pc12, aes(x=PC1, y=PC2))+geom_point(aes(color=factor(grp)), size=r)+labs(title="PCA visualiztion")
ggplot2::aes(x=PC1, y=PC2)
dev.off()
8. 对RNA_seq_2018_08_16/2020_12_17绘制热图
# heatmap ----------------------------------------------------------------------
#heatmap绘制时,需要使用gene_symbol和FPKM值
#关于FPKM值的说明
#在绘制heatmap热图时,需要使用差异基因所对应的FPKM值,该值的获取有几个办法:
#利用Stringtie的-A参数直接获得
#利用DESeq2的-FPKM参数由count转化获得
#利用ballgown包进行转化获得
#本次采用StringTie的-A参数直接获得
#关于gene_symbol的说明
#对于绘制heatmap图的gene,使用具有差异性的基因进行绘图
#利用之前DESeq2分析得到的abs(log2FoldChange)>=1的基因来做图
#需要根据"significant_different_genes"来从包含FPKM值的文件中将其提取出来,使用到了match()函数
#绘图前的准备工作
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
##将得到的各样本的FPKM值数据保存入heatmap目录
write.table(Scr.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Scr.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_3_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_3_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_4_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_4_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
#对差异基因list进行整理(采用pvalue定义的差异基因进行分析)
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes")
##将各实验组的差异基因导入当前工作环境
group_up <- read.csv("significant_pvalue_different_genes_genecount_up.csv")
group_dn <- read.csv("significant_pvalue_different_genes_genecount_down.csv")
##将各实验组的差异基因list进行提取
group_up_gene <- group_up[,1]
group_dn_gene <- group_dn[,1]
##查看各差异基因名信息
View(group_up_gene)
View(group_dn_gene)
#利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##对group_up各样品进行处理
Scr_group_up_row.NO <- c(match(group_up_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_up_row.NO <- c(match(group_up_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_up_row.NO <- c(match(group_up_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_2.FPKM$Gene.ID))
##对group_dn各样品进行处理
Scr_group_dn_row.NO <- c(match(group_dn_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##对group_up各样品的FPKM值进行提取
Scr_group_up_gene_FPKM <- Scr.FPKM[Scr_group_up_row.NO ,]
shGAS_3_1_group_up_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_up_row.NO ,]
shGAS_4_1_group_up_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_up_row.NO ,]
shGAS_WT_1_group_up_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_up_row.NO ,]
shGAS_WT_2_group_up_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_up_row.NO ,]
shGAS_W93A_1_group_up_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_up_row.NO ,]
shGAS_W93A_2_group_up_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_up_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_up_gene_FPKM)
View(shGAS_3_1_group_up_gene_FPKM)
View(shGAS_4_1_group_up_gene_FPKM)
View(shGAS_WT_1_group_up_gene_FPKM)
View(shGAS_WT_2_group_up_gene_FPKM)
View(shGAS_W93A_1_group_up_gene_FPKM)
View(shGAS_W93A_2_group_up_gene_FPKM)
##对group_dn各样品的FPKM值进行提取
Scr_group_dn_gene_FPKM <- Scr.FPKM[Scr_group_dn_row.NO ,]
shGAS_3_1_group_dn_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_dn_row.NO ,]
shGAS_4_1_group_dn_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_dn_row.NO ,]
shGAS_WT_1_group_dn_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_dn_row.NO ,]
shGAS_WT_2_group_dn_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_dn_row.NO ,]
shGAS_W93A_1_group_dn_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_dn_row.NO ,]
shGAS_W93A_2_group_dn_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_dn_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_dn_gene_FPKM)
View(shGAS_3_1_group_dn_gene_FPKM)
View(shGAS_4_1_group_dn_gene_FPKM)
View(shGAS_WT_1_group_dn_gene_FPKM)
View(shGAS_WT_2_group_dn_gene_FPKM)
View(shGAS_W93A_1_group_dn_gene_FPKM)
View(shGAS_W93A_2_group_dn_gene_FPKM)
#利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
##对group_up各样品的FPKM值进行合并
group_up_gene_FPKM <- merge(Scr_group_up_gene_FPKM, merge(merge(merge(shGAS_3_1_group_up_gene_FPKM, shGAS_4_1_group_up_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_up_gene_FPKM, shGAS_WT_2_group_up_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_up_gene_FPKM, shGAS_W93A_2_group_up_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##对group_dn各样品的FPKM值进行合并
group_dn_gene_FPKM <- merge(Scr_group_dn_gene_FPKM, merge(merge(merge(shGAS_3_1_group_dn_gene_FPKM, shGAS_4_1_group_dn_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_dn_gene_FPKM, shGAS_WT_2_group_dn_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_dn_gene_FPKM, shGAS_W93A_2_group_dn_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##查看合并结果,确认
View(group_up_gene_FPKM)
View(group_dn_gene_FPKM)
#将各实验组差异基因对应的FPKM数据保存至heatmap文件中
write.table(group_up_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_up_gene_FPKM", row.names = FALSE)
write.table(group_dn_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_dn_gene_FPKM", row.names = FALSE)
#开始绘制heatmap图啦啦啦啦啦啦啦啦啦
#代码参考网站:https://www.jianshu.com/p/d86e4afe1065
#安装包(作者说这种方式下载的pheatmap包版本更新一些)
#install.packages('devtools')
#library(devtools)
#install_github("raivokolde/pheatmap")
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/")
#加载包
library(RColorBrewer)#设置颜色用的
library(pheatmap)
#设置配色方案
cc = colorRampPalette(rev(brewer.pal(n=7, name="RdYlBu"))) #Rd=red Yl=yellow Bu=blue
#读入文件,如果确实过多,会无法聚类,最好保证没有缺失,或将缺失替换为0
Group_up<-read.table(file = "group_up_gene_FPKM",row.names = 1,header = T,check.names = F)
Group_dn<-read.table(file = "group_dn_gene_FPKM",row.names = 1,header = T,check.names = F)
#如果矩阵内容是fpkm表达量,一般取log10(fpkm+1)绘图
Group_up=log2(Group_up+1)
Group_dn=log2(Group_dn+1)
#pheatmap参数解释:
#第一个参数是需要用pheatmap画图的数据
#color: 设置颜色。如果想画得更精细一些,可以取cc(1000)
#main: 标题,会显示在最上面
#fontsize: row的字体大小
#scale: 是否归一化为正态分布,可选row,column,none。一般对row进行归一化的情况比较多,column较少。
#border_color: 是否显示边框及边框的颜色,NA不显示, red显示红色。支持简单的颜色单词
#na_col: 设置缺失值的颜色,支持简单颜色单词,一般设置为灰色就满好识别的。
#cluster_rows & cluster_cols: 设置是否对行进行聚类,这个就见仁见智,看你的实际需求了。当缺失值较多的时候是无法进行聚类的。一个解决办法是读取数据的时候不设置缺失值。
#show_rownames & show_colnames: 是否显示行/列的名称
#treeheight_row & treeheight_col: 当前面设置了聚类之后,两边会出现聚类的树,这个参数是设置树的高度的。
#cellheight & cellwidth: 设置每个各自的宽度和高度。有的时候不设置这两个值画出来的树容易放飞自我????
#cutree_row & cutree_col: 是否根据聚类情况把树切开,可以设置切开的份数。
#display_numbers: 设置是否显示每个单元格的值。这个也是个人喜好及文章需求。
#legend: 设置是否显示旁边的bar状图例,emmmm好像还没碰到说不要那个玩意儿的情况。。
#filename: 设置输出文件的名字。可以设置的文件类型有:pdf,png,jpg,tiff,bmp
#绘图group_up
heatmap=pheatmap(Group_up,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_up.tiff")
#绘图group_dn
heatmap=pheatmap(Group_dn,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_dn.tiff")
#采用padj定义的基因进行热图绘制-------------------------------------------------------------------------------------------------------------------------
# heatmap ----------------------------------------------------------------------
#heatmap绘制时,需要使用gene_symbol和FPKM值
#关于FPKM值的说明
#在绘制heatmap热图时,需要使用差异基因所对应的FPKM值,该值的获取有几个办法:
#利用Stringtie的-A参数直接获得
#利用DESeq2的-FPKM参数由count转化获得
#利用ballgown包进行转化获得
#本次采用StringTie的-A参数直接获得
#关于gene_symbol的说明
#对于绘制heatmap图的gene,使用具有差异性的基因进行绘图
#利用之前DESeq2分析得到的abs(log2FoldChange)>=1的基因来做图
#需要根据"significant_different_genes"来从包含FPKM值的文件中将其提取出来,使用到了match()函数
#绘图前的准备工作
#对FPKM数据进行整理
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
##将StringTie分析得到的含有FPKM数据的TAB文件导入当前工作环境中
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/")
Scr.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_scr_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_3_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh3_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_4_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2018_08_16/gene_tab/KD_sh4_1.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/")
shGAS_WT_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_WT_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_WT_2.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_1.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
shGAS_W93A_2.gene.tab <- read.table("G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Rescue_W93A_1.clean.gene.tab", header = TRUE, sep = "\t" , quote = "\"")
##提取指定列的内容
###对数据中的Gene.ID和FPKM两列数据进行提取
Scr.FPKM <- Scr.gene.tab[,c(1,8)]
shGAS_3_1.FPKM <- shGAS_3_1.gene.tab[,c(1,8)]
shGAS_4_1.FPKM <- shGAS_4_1.gene.tab[,c(1,8)]
shGAS_WT_1.FPKM <- shGAS_WT_1.gene.tab[,c(1,8)]
shGAS_WT_2.FPKM <- shGAS_WT_2.gene.tab[,c(1,8)]
shGAS_W93A_1.FPKM <- shGAS_W93A_1.gene.tab[,c(1,8)]
shGAS_W93A_2.FPKM <- shGAS_W93A_2.gene.tab[,c(1,8)]
###重命名指定列
###重命名全部的列是name(data) <- c("NO","name")
###重命名单个列是colnames(data)[2] <- 'newname'
colnames(Scr.FPKM)[2] <-"Scr"
colnames(shGAS_3_1.FPKM)[2] <-"shGAS_3_1"
colnames(shGAS_4_1.FPKM)[2] <-"shGAS_4_1"
colnames(shGAS_WT_1.FPKM)[2] <-"shGAS_WT_1"
colnames(shGAS_WT_2.FPKM)[2] <-"shGAS_WT_2"
colnames(shGAS_W93A_1.FPKM)[2] <-"shGAS_W93A_1"
colnames(shGAS_W93A_2.FPKM)[2] <-"shGAS_W93A_2"
##将得到的各样本的FPKM值数据保存入heatmap目录
write.table(Scr.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/Scr.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_3_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_3_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_4_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_4_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_WT_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_WT_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_1.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_1.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
write.table(shGAS_W93A_2.FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/gene_tab/shGAS_W93A_2.FPKM", row.names = FALSE, quote = FALSE, sep = "\t")
#对差异基因list进行整理(采用padj定义的差异基因进行分析)
##设置工作目录
setwd("G:/dongfeng/RNA-seq/2018_08_16/Rtreatment/significant_different_genes")
##将各实验组的差异基因导入当前工作环境
group_up <- read.csv("significant_padj_different_genes_genecount_up.csv")
group_dn <- read.csv("significant_padj_different_genes_genecount_down.csv")
##将各实验组的差异基因list进行提取
group_up_gene <- group_up[,1]
group_dn_gene <- group_dn[,1]
##查看各差异基因名信息
View(group_up_gene)
View(group_dn_gene)
#利用match函数对差异基因List信息(小文件)和FPKM值信息(大文件)进行提取
##利用match函数提取各样品中差异基因所在的行数并重新命名为row.NO文件
##match(x,y)函数输出结果:x向量在y向量中所处的位置,x向量元素不存在y向量中的返回NA
##match(x, table$i)函数输出结果:返回x向量在table中$i列中所处的位置
##对group_up各样品进行处理
Scr_group_up_row.NO <- c(match(group_up_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_up_row.NO <- c(match(group_up_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_up_row.NO <- c(match(group_up_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_up_row.NO <- c(match(group_up_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_up_row.NO <- c(match(group_up_gene, shGAS_W93A_2.FPKM$Gene.ID))
##对group_dn各样品进行处理
Scr_group_dn_row.NO <- c(match(group_dn_gene, Scr.FPKM$Gene.ID))
shGAS_3_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_3_1.FPKM$Gene.ID))
shGAS_4_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_4_1.FPKM$Gene.ID))
shGAS_WT_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_1.FPKM$Gene.ID))
shGAS_WT_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_WT_2.FPKM$Gene.ID))
shGAS_W93A_1_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_1.FPKM$Gene.ID))
shGAS_W93A_2_group_dn_row.NO <- c(match(group_dn_gene, shGAS_W93A_2.FPKM$Gene.ID))
#根据以上行数,对各样品的FPKM值进行提取
##对group_up各样品的FPKM值进行提取
Scr_group_up_gene_FPKM <- Scr.FPKM[Scr_group_up_row.NO ,]
shGAS_3_1_group_up_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_up_row.NO ,]
shGAS_4_1_group_up_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_up_row.NO ,]
shGAS_WT_1_group_up_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_up_row.NO ,]
shGAS_WT_2_group_up_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_up_row.NO ,]
shGAS_W93A_1_group_up_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_up_row.NO ,]
shGAS_W93A_2_group_up_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_up_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_up_gene_FPKM)
View(shGAS_3_1_group_up_gene_FPKM)
View(shGAS_4_1_group_up_gene_FPKM)
View(shGAS_WT_1_group_up_gene_FPKM)
View(shGAS_WT_2_group_up_gene_FPKM)
View(shGAS_W93A_1_group_up_gene_FPKM)
View(shGAS_W93A_2_group_up_gene_FPKM)
##对group_dn各样品的FPKM值进行提取
Scr_group_dn_gene_FPKM <- Scr.FPKM[Scr_group_dn_row.NO ,]
shGAS_3_1_group_dn_gene_FPKM <- shGAS_3_1.FPKM[shGAS_3_1_group_dn_row.NO ,]
shGAS_4_1_group_dn_gene_FPKM <- shGAS_4_1.FPKM[shGAS_4_1_group_dn_row.NO ,]
shGAS_WT_1_group_dn_gene_FPKM <- shGAS_WT_1.FPKM[shGAS_WT_1_group_dn_row.NO ,]
shGAS_WT_2_group_dn_gene_FPKM <- shGAS_WT_2.FPKM[shGAS_WT_2_group_dn_row.NO ,]
shGAS_W93A_1_group_dn_gene_FPKM <- shGAS_W93A_1.FPKM[shGAS_W93A_1_group_dn_row.NO ,]
shGAS_W93A_2_group_dn_gene_FPKM <- shGAS_W93A_2.FPKM[shGAS_W93A_2_group_dn_row.NO ,]
###查看提取结果,确认各样品行数和基因名一致性
View(Scr_group_dn_gene_FPKM)
View(shGAS_3_1_group_dn_gene_FPKM)
View(shGAS_4_1_group_dn_gene_FPKM)
View(shGAS_WT_1_group_dn_gene_FPKM)
View(shGAS_WT_2_group_dn_gene_FPKM)
View(shGAS_W93A_1_group_dn_gene_FPKM)
View(shGAS_W93A_2_group_dn_gene_FPKM)
#利用merge函数对各组实验的FPKM值进行合并
##merge(x,y, by="")
##对group_up各样品的FPKM值进行合并
group_up_gene_FPKM <- merge(Scr_group_up_gene_FPKM, merge(merge(merge(shGAS_3_1_group_up_gene_FPKM, shGAS_4_1_group_up_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_up_gene_FPKM, shGAS_WT_2_group_up_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_up_gene_FPKM, shGAS_W93A_2_group_up_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##对group_dn各样品的FPKM值进行合并
group_dn_gene_FPKM <- merge(Scr_group_dn_gene_FPKM, merge(merge(merge(shGAS_3_1_group_dn_gene_FPKM, shGAS_4_1_group_dn_gene_FPKM,by="Gene.ID"), merge(shGAS_WT_1_group_dn_gene_FPKM, shGAS_WT_2_group_dn_gene_FPKM,by="Gene.ID"), by="Gene.ID"),merge(shGAS_W93A_1_group_dn_gene_FPKM, shGAS_W93A_2_group_dn_gene_FPKM,by="Gene.ID"),by="Gene.ID"),by="Gene.ID")
##查看合并结果,确认
View(group_up_gene_FPKM)
View(group_dn_gene_FPKM)
#将各实验组差异基因对应的FPKM数据保存至heatmap文件中
write.table(group_up_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_up_gene_padj_FPKM", row.names = FALSE)
write.table(group_dn_gene_FPKM, file = "G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/group_dn_gene_padj_FPKM", row.names = FALSE)
#开始绘制heatmap图啦啦啦啦啦啦啦啦啦
#代码参考网站:https://www.jianshu.com/p/d86e4afe1065
#安装包(作者说这种方式下载的pheatmap包版本更新一些)
#install.packages('devtools')
#library(devtools)
#install_github("raivokolde/pheatmap")
#清空环境变量
rm(list=ls())
#获取当前工作目录
getwd()
#设置工作目录
setwd("G:/dongfeng/RNA-seq/2020_12_17/Rtreatment/heatmap/")
#加载包
library(RColorBrewer)#设置颜色用的
library(pheatmap)
#设置配色方案
cc = colorRampPalette(rev(brewer.pal(n=7, name="RdYlBu"))) #Rd=red Yl=yellow Bu=blue
#读入文件,如果确实过多,会无法聚类,最好保证没有缺失,或将缺失替换为0
Group_up<-read.table(file = "group_up_gene_padj_FPKM",row.names = 1,header = T,check.names = F)
Group_dn<-read.table(file = "group_dn_gene_padj_FPKM",row.names = 1,header = T,check.names = F)
#如果矩阵内容是fpkm表达量,一般取log10(fpkm+1)绘图
Group_up=log2(Group_up+1)
Group_dn=log2(Group_dn+1)
#pheatmap参数解释:
#第一个参数是需要用pheatmap画图的数据
#color: 设置颜色。如果想画得更精细一些,可以取cc(1000)
#main: 标题,会显示在最上面
#fontsize: row的字体大小
#scale: 是否归一化为正态分布,可选row,column,none。一般对row进行归一化的情况比较多,column较少。
#border_color: 是否显示边框及边框的颜色,NA不显示, red显示红色。支持简单的颜色单词
#na_col: 设置缺失值的颜色,支持简单颜色单词,一般设置为灰色就满好识别的。
#cluster_rows & cluster_cols: 设置是否对行进行聚类,这个就见仁见智,看你的实际需求了。当缺失值较多的时候是无法进行聚类的。一个解决办法是读取数据的时候不设置缺失值。
#show_rownames & show_colnames: 是否显示行/列的名称
#treeheight_row & treeheight_col: 当前面设置了聚类之后,两边会出现聚类的树,这个参数是设置树的高度的。
#cellheight & cellwidth: 设置每个各自的宽度和高度。有的时候不设置这两个值画出来的树容易放飞自我????
#cutree_row & cutree_col: 是否根据聚类情况把树切开,可以设置切开的份数。
#display_numbers: 设置是否显示每个单元格的值。这个也是个人喜好及文章需求。
#legend: 设置是否显示旁边的bar状图例,emmmm好像还没碰到说不要那个玩意儿的情况。。
#filename: 设置输出文件的名字。可以设置的文件类型有:pdf,png,jpg,tiff,bmp
#绘图group_up
heatmap=pheatmap(Group_up,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_padj_up.tiff")
#绘图group_dn
heatmap=pheatmap(Group_dn,color = cc(1000),
main=" ",
fontsize = 15,
scale="row",
border_color = NA,
na_col = "grey",
cluster_rows = T,cluster_cols = F,
show_rownames = T,show_colnames = T,
treeheight_row = 30,treeheight_col = 30,
cellheight = 15,cellwidth = 30,
cutree_row=2,cutree_col=2,
display_numbers = F,legend = T,
filename = "Group_padj_dn.tiff")