【心得】| 基于百度paddlepaddle框架的图游走模型学习
引言
我们都知道在数据结构中,图是一种基础且常用的结构。现实世界中许多场景可以抽象为一种图结构,如社交网络,交通网络,电商网站中用户与物品的关系等。
目前提到图算法一般指:
1. 经典数据结构与算法层面的:最小生成树(Prim,Kruskal,...),最短路(Dijkstra,Floyed,...),拓扑排序,关键路径等
2. 概率图模型,涉及图的表示,推断和学习,详细可以参考Koller的书或者公开课
3. 图神经网络,主要包括Graph Embedding(基于随机游走)和Graph CNN(基于邻居汇聚)两部分。
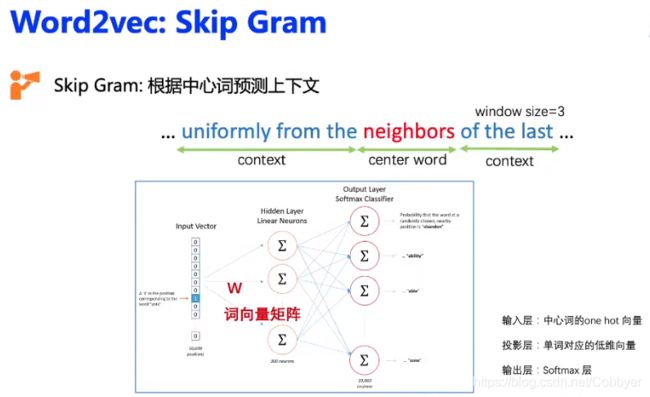
图游走算法最先参考的是NLP的Word2vec模型,Word2vec模型的其中一种方法是Skip Gram,即根据中心词预测上下文,之后通过负采样的方式进行优化。将Word2vec的思想和图结合起来就会得到了图游走类算法。
DeepWalk 算法原理
DeepWalk的思想类似word2vec,使用图中节点与节点的共现关系来学习节点的向量表示。那么关键的问题就是如何来描述节点与节点的共现关系,DeepWalk给出的方法是使用随机游走(RandomWalk)的方式在图中进行节点采样。RandomWalk是一种可重复访问已访问节点的深度优先遍历算法。给定当前访问起始节点,从其邻居中随机采样节点作为下一个访问节点,重复此过程,直到访问序列长度满足预设条件。获取足够数量的节点访问序列后,使用skip-gram model 进行向量学习。
DeepWalk算法主要包括两个步骤,第一步为随机游走采样节点序列,第二步为使用skip-gram modelword2vec学习表达向量。
①构建同构网络,从网络中的每个节点开始分别进行Random Walk 采样,得到局部相关联的训练数据;
②对采样数据进行SkipGram训练,将离散的网络节点表示成向量化,最大化节点共现,使用Hierarchical Softmax来做超大规模分类的分类器
使用随机游走有两个好处:
- 并行化,随机游走是局部的,对于一个大的网络来说,可以同时在不同的顶点开始进行一定长度的随机游走,多个随机游走同时进行,可以减少采样的时间。
- 适应性,可以适应网络局部的变化。网络的演化通常是局部的点和边的变化,这样的变化只会对部分随机游走路径产生影响,因此在网络的演化过程中不需要每一次都重新计算整个网络的随机游走。
DeepWalk存在的问题是比较简单直接,而图结构往往是一个复杂结构,需要考虑很多因素,在深度优先搜索方法之外,还有广度优先搜索,结合以上两种方式可以更好的探索图模型,即node2vec。node2vec和DeepWalk相比主要修改的是转移概率分布,不同于随机游走相邻节点转移的概率相同,node2vec考虑了边的权值和节点之间的距离,具体如下:
基于百度paddlepaddle框架实现图结构及图的游走模型
1. DeepWalk采样算法
对于给定的节点,DeepWalk会等概率的选取下一个相邻节点加入路径,直至达到最大路径长度,或者没有下一个节点可选。
from pgl.graph import Graph
import numpy as np
class UserDefGraph(Graph):
def random_walk(self, nodes, walk_len):
"""
输入:nodes - 当前节点id list (batch_size,)
walk_len - 最大路径长度 int
输出:以当前节点为起点得到的路径 list (batch_size, walk_len)
用到的函数
1. self.successor(nodes)
描述:获取当前节点的下一个相邻节点id列表
输入:nodes - list (batch_size,)
输出:succ_nodes - list of list ((num_successors_i,) for i in range(batch_size))
2. self.outdegree(nodes)
描述:获取当前节点的出度
输入:nodes - list (batch_size,)
输出:out_degrees - list (batch_size,)
"""
walks = [[node] for node in nodes]
walks_ids = np.arange(0, len(nodes))
cur_nodes = np.array(nodes)
for l in range(walk_len):
"""选取有下一个节点的路径继续采样,否则结束"""
outdegree = self.outdegree(cur_nodes)
walk_mask = (outdegree != 0)
if not np.any(walk_mask):
break
cur_nodes = cur_nodes[walk_mask]
walks_ids = walks_ids[walk_mask]
outdegree = outdegree[walk_mask]
succ_nodes = self.successor(cur_nodes)
sample_index = np.floor(np.random.rand(outdegree.shape[0])*outdegree).astype("int64")
next_nodes = []
for s, ind, walk_id in zip(succ_nodes, sample_index, walks_ids):
walks[walk_id].append(s[ind])
next_nodes.append(s[ind])
cur_nodes = np.array(next_nodes)
return walks2. SkipGram模型训练
在得到节点路径后,node2vec会使用SkipGram模型学习节点表示,给定中心节点,预测局部路径中还有哪些节点。模型中用了negative sampling来降低计算量。
import paddle.fluid.layers as l
def userdef_loss(embed_src, weight_pos, weight_negs):
"""
输入:embed_src - 中心节点向量 list (batch_size, 1, embed_size)
weight_pos - 标签节点向量 list (batch_size, 1, embed_size)
weight_negs - 负样本节点向量 list (batch_size, neg_num, embed_size)
输出:loss - 正负样本的交叉熵 float
"""
pos_logits = l.matmul(
embed_src, weight_pos, transpose_y=True) # [batch_size, 1, 1]
neg_logits = l.matmul(
embed_src, weight_negs, transpose_y=True) # [batch_size, 1, neg_num]
ones_label = pos_logits * 0. + 1.
ones_label.stop_gradient = True
pos_loss = l.sigmoid_cross_entropy_with_logits(pos_logits, ones_label)
zeros_label = neg_logits * 0.
zeros_label.stop_gradient = True
neg_loss = l.sigmoid_cross_entropy_with_logits(neg_logits, zeros_label)
loss = (l.reduce_mean(pos_loss) + l.reduce_mean(neg_loss)) / 2
return loss3. Node2Vec采样算法
Node2Vec会根据与上个节点的距离按不同概率采样得到当前节点的下一个节点。
import numpy as np
def node2vec_sample(succ, prev_succ, prev_node, p, q):
"""
输入:succ - 当前节点的下一个相邻节点id列表 list (num_neighbors,)
prev_succ - 前一个节点的下一个相邻节点id列表 list (num_neighbors,)
prev_node - 前一个节点id int
p - 控制回到上一节点的概率 float
q - 控制偏向DFS还是BFS float
输出:下一个节点id int
"""
succ_len = len(succ)
prev_succ_len = len(prev_succ)
prev_succ_set = np.asarray([])
for i in range(prev_succ_len):
prev_succ_set = np.append(prev_succ_set,prev_succ[i])
# 概率参数信息
probs = []
prob = 0
prob_sum = 0.
for i in range(succ_len):
if succ[i] == prev_node:
prob = 1. / p
elif np.where(prev_succ_set==succ[i]):
prob = 1.
elif np.where(prev_succ_set!=succ[i]):
prob = 1. / q
else:
prob = 0.
probs.append(prob)
prob_sum += prob
RAND_MAX = 65535
rand_num = float(np.random.randint(0, RAND_MAX+1)) / RAND_MAX * prob_sum
sampled_succ = 0.
for i in range(succ_len):
rand_num -= probs[i]
if rand_num <= 0:
sampled_succ = succ[i]
return sampled_succ 此处给出的是为了方便实现原始公示的代码,原版PGL代码在此基础上进行了优化,提供PGL源码中的node2vec代码供参考,原版PGL代码在:https://github.com/PaddlePaddle/PGL,PGL依托于百度paddlepaddle,是百度开发出的优秀深度学习框架,亲测好用。
# Copyright (c) 2019 PaddlePaddle Authors. All Rights Reserved
#
# Licensed under the Apache License, Version 2.0 (the "License");
# you may not use this file except in compliance with the License.
# You may obtain a copy of the License at
#
# http://www.apache.org/licenses/LICENSE-2.0
#
# Unless required by applicable law or agreed to in writing, software
# distributed under the License is distributed on an "AS IS" BASIS,
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# See the License for the specific language governing permissions and
# limitations under the License.
import argparse
import time
import math
import os
import io
from multiprocessing import Pool
import glob
import numpy as np
import sklearn.metrics
from sklearn.metrics import f1_score
import pgl
from pgl import data_loader
from pgl.utils import op
from pgl.utils.logger import log
import paddle.fluid as fluid
import paddle.fluid.layers as l
def load(name):
if name == "BlogCatalog":
dataset = data_loader.BlogCatalogDataset()
elif name == "ArXiv":
dataset = data_loader.ArXivDataset()
else:
raise ValueError(name + " dataset doesn't exists")
return dataset
def node2vec_model(graph, hidden_size=16, neg_num=5):
pyreader = l.py_reader(
capacity=70,
shapes=[[-1, 1, 1], [-1, 1, 1], [-1, neg_num, 1]],
dtypes=['int64', 'int64', 'int64'],
lod_levels=[0, 0, 0],
name='train',
use_double_buffer=True)
embed_init = fluid.initializer.UniformInitializer(low=-1.0, high=1.0)
weight_init = fluid.initializer.TruncatedNormal(scale=1.0 /
math.sqrt(hidden_size))
src, pos, negs = l.read_file(pyreader)
embed_src = l.embedding(
input=src,
size=[graph.num_nodes, hidden_size],
param_attr=fluid.ParamAttr(
name='content', initializer=embed_init))
weight_pos = l.embedding(
input=pos,
size=[graph.num_nodes, hidden_size],
param_attr=fluid.ParamAttr(
name='weight', initializer=weight_init))
weight_negs = l.embedding(
input=negs,
size=[graph.num_nodes, hidden_size],
param_attr=fluid.ParamAttr(
name='weight', initializer=weight_init))
pos_logits = l.matmul(
embed_src, weight_pos, transpose_y=True) # [batch_size, 1, 1]
neg_logits = l.matmul(
embed_src, weight_negs, transpose_y=True) # [batch_size, 1, neg_num]
ones_label = pos_logits * 0. + 1.
ones_label.stop_gradient = True
pos_loss = l.sigmoid_cross_entropy_with_logits(pos_logits, ones_label)
zeros_label = neg_logits * 0.
zeros_label.stop_gradient = True
neg_loss = l.sigmoid_cross_entropy_with_logits(neg_logits, zeros_label)
loss = (l.reduce_mean(pos_loss) + l.reduce_mean(neg_loss)) / 2
return pyreader, loss
def gen_pair(walks, left_win_size=2, right_win_size=2):
src = []
pos = []
for walk in walks:
for left_offset in range(1, left_win_size + 1):
src.extend(walk[left_offset:])
pos.extend(walk[:-left_offset])
for right_offset in range(1, right_win_size + 1):
src.extend(walk[:-right_offset])
pos.extend(walk[right_offset:])
src, pos = np.array(src, dtype=np.int64), np.array(pos, dtype=np.int64)
src, pos = np.expand_dims(src, -1), np.expand_dims(pos, -1)
src, pos = np.expand_dims(src, -1), np.expand_dims(pos, -1)
return src, pos
def node2vec_generator(graph,
batch_size=512,
walk_len=5,
p=0.25,
q=0.25,
win_size=2,
neg_num=5,
epoch=200,
filelist=None):
def walks_generator():
if filelist is not None:
bucket = []
for filename in filelist:
with io.open(filename) as inf:
for line in inf:
walk = [int(x) for x in line.strip('\n').split(' ')]
bucket.append(walk)
if len(bucket) == batch_size:
yield bucket
bucket = []
if len(bucket):
yield bucket
else:
for _ in range(epoch):
for nodes in graph.node_batch_iter(batch_size):
walks = graph.node2vec_random_walk(nodes, walk_len, p, q)
yield walks
def wrapper():
for walks in walks_generator():
src, pos = gen_pair(walks, win_size, win_size)
if src.shape[0] == 0:
continue
negs = graph.sample_nodes([len(src), neg_num, 1]).astype(np.int64)
yield [src, pos, negs]
return wrapper
def process(args):
idx, graph, save_path, epoch, batch_size, walk_len, p, q, seed = args
with open('%s/%s' % (save_path, idx), 'w') as outf:
for _ in range(epoch):
np.random.seed(seed)
for nodes in graph.node_batch_iter(batch_size):
walks = graph.node2vec_random_walk(nodes, walk_len, p, q)
for walk in walks:
outf.write(' '.join([str(token) for token in walk]) + '\n')
def main(args):
hidden_size = args.hidden_size
neg_num = args.neg_num
epoch = args.epoch
p = args.p
q = args.q
save_path = args.save_path
batch_size = args.batch_size
walk_len = args.walk_len
win_size = args.win_size
if not os.path.isdir(save_path):
os.makedirs(save_path)
dataset = load(args.dataset)
if args.offline_learning:
log.info("Start random walk on disk...")
walk_save_path = os.path.join(save_path, "walks")
if not os.path.isdir(walk_save_path):
os.makedirs(walk_save_path)
pool = Pool(args.processes)
args_list = [(x, dataset.graph, walk_save_path, 1, batch_size,
walk_len, p, q, np.random.randint(2**32))
for x in range(epoch)]
pool.map(process, args_list)
filelist = glob.glob(os.path.join(walk_save_path, "*"))
log.info("Random walk on disk Done.")
else:
filelist = None
train_steps = int(dataset.graph.num_nodes / batch_size) * epoch
place = fluid.CUDAPlace(0) if args.use_cuda else fluid.CPUPlace()
node2vec_prog = fluid.Program()
startup_prog = fluid.Program()
with fluid.program_guard(node2vec_prog, startup_prog):
with fluid.unique_name.guard():
node2vec_pyreader, node2vec_loss = node2vec_model(
dataset.graph, hidden_size=hidden_size, neg_num=neg_num)
lr = l.polynomial_decay(0.025, train_steps, 0.0001)
adam = fluid.optimizer.Adam(lr)
adam.minimize(node2vec_loss)
node2vec_pyreader.decorate_tensor_provider(
node2vec_generator(
dataset.graph,
batch_size=batch_size,
walk_len=walk_len,
win_size=win_size,
epoch=epoch,
neg_num=neg_num,
p=p,
q=q,
filelist=filelist))
node2vec_pyreader.start()
exe = fluid.Executor(place)
exe.run(startup_prog)
prev_time = time.time()
step = 0
while 1:
try:
node2vec_loss_val = exe.run(node2vec_prog,
fetch_list=[node2vec_loss],
return_numpy=True)[0]
cur_time = time.time()
use_time = cur_time - prev_time
prev_time = cur_time
step += 1
log.info("Step %d " % step + "Node2vec Loss: %f " %
node2vec_loss_val + " %f s/step." % use_time)
except fluid.core.EOFException:
node2vec_pyreader.reset()
break
fluid.io.save_persistables(exe,
os.path.join(save_path, "paddle_model"),
node2vec_prog)
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='node2vec')
parser.add_argument(
"--dataset",
type=str,
default="BlogCatalog",
help="dataset (BlogCatalog, ArXiv)")
parser.add_argument("--use_cuda", action='store_true', help="use_cuda")
parser.add_argument(
"--offline_learning", action='store_true', help="use_cuda")
parser.add_argument("--hidden_size", type=int, default=128)
parser.add_argument("--neg_num", type=int, default=20)
parser.add_argument("--epoch", type=int, default=100)
parser.add_argument("--batch_size", type=int, default=1024)
parser.add_argument("--walk_len", type=int, default=40)
parser.add_argument("--win_size", type=int, default=10)
parser.add_argument("--p", type=float, default=0.25)
parser.add_argument("--q", type=float, default=0.25)
parser.add_argument("--save_path", type=str, default="./tmp/node2vec")
parser.add_argument("--processes", type=int, default=10)
args = parser.parse_args()
log.info(args)
main(args)
最后在ArXiv数据集上学习节点表示,预测合作关系的实验结果如下:
总结
本文心得是参加百度图神经网络7日打卡营活动有感而发,在这个活动中进一步加强了自己图神经网络GCN的理论基础,并且学习了在NLP中广泛使用的图游走算法。为后面的图学习科研之路打下更深的理论基础。本人研究方向是计算机视觉方向,通过这几天的打卡营活动,感悟颇多,图游走算法在NLP中已经取得了优秀的成果,相信在CV中也能够取得优异的表现。
同时,本人也是第二次参加百度的打卡营活动,这活动在AI Studio平台上进行,代码编写及模型调参都非常方便。非常适合科研er进行学习使用,希望百度的paddlepaddle框架能够越来越优秀,和Pytorch及Tf框架平分天下,期待国产崛起。欢迎大家给百度paddlepaddle的PGL多多star哦!地址在这:https://github.com/PaddlePaddle/PGL。大家可以在百度AI Studio学习平台上学习更多的AI知识:https://aistudio.baidu.com/aistudio/index