R语言|绘制森林图
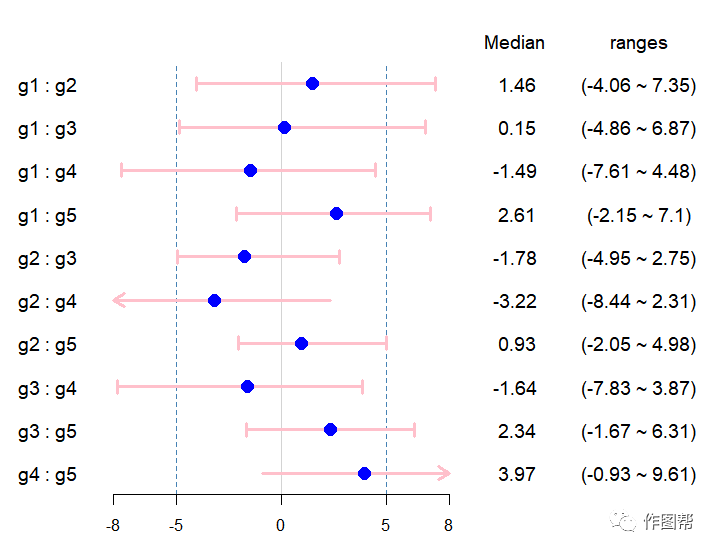
森林图指的是对不同研究提出同一研究问题,用一系列轴来呈现转换为同一统计量的研究结果。除了meta 分析外,森林图还广泛应用于观察性研究和临床试验,例如风险分析/生存分析等结果。最大的优势是简单直观地展示单一研究和汇总研究。今天小编就用forestplot包绘制森林图,仅供参考哦~
代码如下:
1.安装并调用forestplot包
BiocManager::install("forestplot")
library(forestplot)
2.生成模拟数据,共5个模型
g1 <- rnorm(100, 8, 4)
g2 <- rnorm(100, 6, 3)
g3 <- rnorm(100, 7, 3.5)
g4 <- rnorm(100, 9, 4.5)
g5 <- rnorm(100, 5, 2.5)
Data <- data.frame(g1=g1,g2=g2,g3=g3,g4=g4,g5=g5)
head(Data)
3.计算各个模型之间的15%分位数,中位数,85%分位数,同时保留两位有效数字
Data_difference<-apply(Data(round(quantile(x,c(0.15,0.5,0.85)),digits=2))
4.设置行名
rownames(Data_difference)<-c("down","median","up")
5.各个模型间的比较
comparison_name <- c("g1 : g2","g1 : g3","g1 : g4", "g1 : g5",
"g2 : g3","g2 : g4","g2 : g5",
"g3 : g4","g3 : g5","g4 : g5")
6.误差条的差值计算
median <- Data_t$median #差值的中值
ranges <- paste("(", Data_t$down, " ~ ", Data_t$up, ")", sep = "") #差值的四分距的范围
7.误差条的显示数据
Data_str<- data.frame(pairs_na=comparison_name,pairs_median=median,pairs_CI=ranges)
Data_str <- as.matrix(Data_str) #类型转为矩阵
Data_str <- rbind(c(NA,"Median","ranges"),Data_str) #第一行表示指标说明,NA表示不显示
8.数据可视化
forestplot(Data_str,#显示的文本
c(NA,Data_t$median), #误差条的均值(此处为差值的中值)
c(NA,Data_t$down), #误差条的下界(此处为差值的15%分位数)
c(NA,Data_t$up), #误差条的上界(此处为差值的85%分位数)
zero = 0,#显示y=0的垂直线
xlog=FALSE, #x轴的坐标不取对数
grid = structure(c(-5, 5), gp = gpar(col = "steelblue", lty = 2)),#grid设置80%和120%的线条
clip = c(1, 1.5),#设置坐标轴
xticks = c(-8,-5,0,5,8),#X坐标轴的标度
txt_gp = fpTxtGp(ticks = gpar(cex = 1), xlab = gpar(cex = 1.5), cex = 1.2),#文本大小设置
boxsize = 0.3,#误差条中的圆心点大小
graph.pos = 2, # 控制图形输出再第几列
line.margin = 0.1,
fn.ci_norm = fpDrawCircleCI,#误差条中间图形的形状
col=fpColors(line = "pink", #误差条的线的颜色
box="blue"), #误差条的圆心点的颜色
lty.ci = 7, # 误差条的线的线型
lwd.ci = 3, # 误差条的线的宽度
ci.vertices.height = 0.15# 误差条末端的长度
)