异常检测——孤立森林IsolationForest、PCA+马氏距离
文章目录
- 一、孤立森林
-
- 1、孤立森林理论简介
- 2、应用:
- 3、注意:
- 4、关键参数
- 5、python源码
- 二、PCA+马氏距离
-
- 1、原始数据
- 2、处理思路
- 3、python源码
其他方法还有KNN,聚类,暂不考虑。
一、孤立森林
开始是想要识别出变压器的异常运行状态的,确实可以挖掘到一些离群点。后来发现也可以用于机器学习初期的数据处理,清洗掉异常点。
1、孤立森林理论简介
孤立森林理论简介,和参数说明
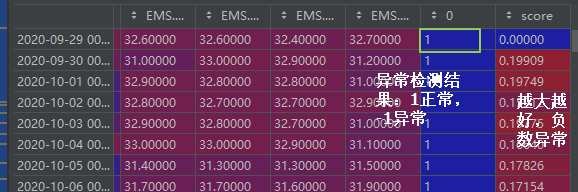
理解:最早被树分离出去(树的长度最短)的数据点,可能为异常点。
2、应用:
- 可以无监督学习,检测异常样本。
- 回归问题中,可以对输出进行异常检测,并去掉这些异常数据,从而提高预测准确率
3、注意:
不需要标准化,不需要PCA降维
4、关键参数
- max_samples=30 估计器的数量,(默认值= 100)
- random_state=rng, rng = np.random.RandomState(30)保证代码的可复现性,便于调试
- contamination=0.1 异常样本占总样本的比例为0.1
5、python源码
import requests
import matplotlib.pyplot as plt
from sklearn.ensemble import IsolationForest
import numpy as np
from sklearn import preprocessing
from sklearn.decomposition import PCA
BIGDATA_DOMAIN = 'http://bigdata-platapi.fnwintranet.com'
BIGDATA_USERKEY = "a95c34cf34deb5a2d0af84f3aea2a616_algorithm-engine-flask"
# "EMS.HZ",
EQUIP_MK_NAME = [
"EMS.Pa", "EMS.Pb", "EMS.Pc", "EMS.P", "EMS.S", "EMS.Q",
"EMS.Ua", "EMS.Ub", "EMS.Uc", "EMS.Uac", "EMS.Ubc", "EMS.Uab",
"EMS.Ia", "EMS.Ib", "EMS.Ic",
"EMS.COSa", "EMS.COSb", "EMS.COSc", "EMS.COS",
"EMS.CHDphAT", "EMS.CHDphBT", "EMS.CHDphCT",
"EMS.VHDphAT", "EMS.VHDphBT", "EMS.VHDphCT", "EMS.VdisPer", "EMS.VHDlineBC", "EMS.VHDlineAC",
"EMS.VHDlineAB",
"EMS.HZ",
"EMS.TphC", "EMS.TphA", "EMS.TphB", "EMS.TphN"
]
def get_iv_data(startTime, endTime, equip_id, station_id, equip_mk, EQUIP_MK_NAME):
tags = {
"equipID": equip_id,
"equipMK": equip_mk,
"staId": station_id
}
d = {
"dataSource": "EMS",
"isClean": False,
"listQueries": [
{
"aggregator": "first",
"downsample": "1d-first-null",
"explicitTags": True,
"metric": i,
"tags": tags
} for i in EQUIP_MK_NAME],
"startTime": startTime,
"endTime": endTime,
"userKey": BIGDATA_USERKEY
}
url = BIGDATA_DOMAIN + '/internal/bigdata/time_series/get_history'
r = requests.post(url, json=d)
return r.json()
import time
import pandas as pd
def iv_data_process(iv_data, EQUIP_MK_NAME):
l_data = [] # 测点值Series列表
l_name = []
for i in range(len(iv_data['data'])):
data = pd.Series(iv_data['data'][i]['dps'])
name = iv_data['data'][i]['metric']
l_data.append(data)
l_name.append(name)
data = pd.concat(l_data, axis=1)
data.columns = l_name
data.index = map(lambda x: time.strftime("%Y-%m-%d %H:%M:%S", time.localtime(int(x))), data.index) # 将时间戳变为datetime
data.sort_index(inplace=True) # 按照index排序
data = data.dropna()
return data
def my_isolationForest(X_train, X_test):
# fit the model
rng = np.random.RandomState(30)
clf = IsolationForest(max_samples=30,
random_state=rng, contamination=0)
clf.fit(X_train)
y_pred_train = clf.predict(X_train)
y_pred_test = clf.predict(X_test)
y_pred_test_decision_function = clf.decision_function(X_test)
y_pred_train_decision_function = clf.decision_function(X_train)
return y_pred_train, y_pred_test, clf, y_pred_test_decision_function, y_pred_train_decision_function
# "IsolationForest"
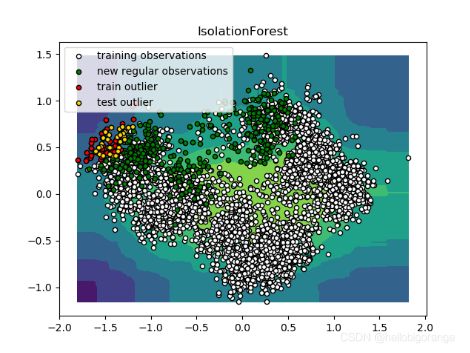
def plot_visualization(clf, title, X_train, X_test, EQUIP_MK_NAME):
xx, yy = np.meshgrid(np.linspace(data[EQUIP_MK_NAME[0]].min(), data[EQUIP_MK_NAME[0]].max(), 100),
np.linspace(data[EQUIP_MK_NAME[1]].min(), data[EQUIP_MK_NAME[1]].max(), 100))
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.title(title)
plt.contourf(xx, yy, Z, camp=plt.cm.Blues_r)
b1 = plt.scatter(X_train[:, 0], X_train[:, 1], c='white',
s=20, edgecolor='k')
b2 = plt.scatter(X_test[:, 0], X_test[:, 1], c='green',
s=20, edgecolor='k')
for i in range(len(X_train)):
if y_pred_train[i] == -1:
b3 = plt.scatter(X_train[i, 0], X_train[i, 1], c='red',
s=20, edgecolor='k')
for i in range(len(X_test)):
if y_pred_test[i] == -1:
b4 = plt.scatter(X_test[i, 0], X_test[i, 1], c='gold',
s=20, edgecolor='k')
plt.axis('tight')
plt.legend([b1, b2],
["training observations",
"new regular observations", "train outlier", "test outlier"])
plt.show()
return Z, np.c_[xx.ravel(), yy.ravel()]
if __name__ == '__main__':
param = {"equipID": "METE01",
"equipMK": "METE",
"staId": "PARK801_EMS01",
"startTime": "2020-09-01 00:00:00",
"endTime": "2021-08-20 23:59:00",
}
equipID = param['equipID']
equipMK = param['equipMK']
staId = param['staId']
startTime = param['startTime']
endTime = param['endTime']
iv_data = get_iv_data(startTime, endTime, equipID, staId, equipMK, EQUIP_MK_NAME)
data = iv_data_process(iv_data, EQUIP_MK_NAME)
# data = normalize_data(data)
# data = pca_process_data(data)
# # 训练集和测试集
X_train = data.loc[startTime:"2021-08-18 00:00:00"].values
X_test = data.loc["2021-08-18 00:15:00":endTime].values
y_pred_train, y_pred_test, clf, y_pred_test_decision_function, y_pred_train_decision_function = my_isolationForest(
X_train, X_test)
y_result = np.concatenate((y_pred_train.reshape(1, len(y_pred_train)), y_pred_test.reshape(1, len(y_pred_test))),
axis=1)
y_result = pd.DataFrame(y_result.reshape(len(y_result[0]), 1), index=data.index)
X_test_data = pd.merge(data, y_result, left_index=True, right_index=True, how='outer')
# # 将输出结果和输入特征进行拼接
# title = "IsolationForest"
# Z, Z_feature = plot_visualization(clf, title, X_train, X_test, ['feature_1', 'feature_2'])
# # 创建一个随机数序列,来查看异常检测的效果
# X_test_random = np.random.randint(1, 100, [10, 2]) # 预测结果为[-1 -1 -1 -1 -1 -1 -1 -1 -1 -1]
# Y_pred_random = clf.predict(X_test_random)
# 拼接上决策得分
y_score = np.concatenate((y_pred_train_decision_function.reshape(1, len(y_pred_train_decision_function)),
y_pred_test_decision_function.reshape(1, len(y_pred_test_decision_function))), axis=1)
y_score = pd.DataFrame(y_score.reshape(len(y_score[0]), 1), index=data.index, columns=['score'])
X_test_data = pd.merge(X_test_data, y_score, left_index=True, right_index=True, how='outer')
plt.plot(X_test_data['score'])
plt.show()
二、PCA+马氏距离
属于统计分布假设检验的算法。
参考博客:
1、PCA+马氏距离用于异常诊断(附数据集和源码)
2、马氏距离定义
1、原始数据
2、处理思路
大致思路就是:
-
首先采用前期较短的正常时间段的数据作为训练数据,然后后续的所有数据作为测试数据。
-
对训练数据进行标准化和PCA降维,再对测试数据进行标准化和PCA降维(我程序写的不对,一起标准化和降维了)
归一化后:

PCA降维后:
-
对处理好的训练集求逆协方差阵和均值
均值:

逆协方差阵:
-
计算训练集和测试集的马氏距离
-
查看训练集马氏距离的分布(认为测试集和训练集分布应该是差不多的),正常的阈值可以为正态分布的95%分布以内。假设阈值为threshold

-
验证集的马氏距离>threshold说明为异常点。
3、python源码
# -*- coding: utf-8 -*-
# @Time : 2021/8/30 9:51
# @Author : Orange
# @File : ma_distance_detection.py
from data_process import *
import matplotlib.pyplot as plt
import numpy as np
from global_variable import *
from get_demand_data import *
def is_pos_def(A):
# 判断A是否为正定矩阵
if np.allclose(A, A.T): # 检查A是否为对称矩阵
try:
np.linalg.cholesky(A)
return True
except np.linalg.LinAlgError:
return False
else:
return False
def cov_matrix(data):
# 求协方差阵和逆协方差阵 cov(X, Y) = E(X-EX)(Y-EY)。
covariance_matrix = np.cov(data, rowvar=False)
if is_pos_def(covariance_matrix):
inv_covariance_matrix = np.linalg.inv(covariance_matrix)
if is_pos_def(inv_covariance_matrix):
return covariance_matrix, inv_covariance_matrix
else:
print("Error: Inverse of Covariance Matrix is not positive definite!")
else:
print("Error: Covariance Matrix is not positive definite!")
def MahalanobisDist(inv_cov_matrix, mean_distr, data, verbose=False):
# 计算马氏距离
inv_covariance_matrix = inv_cov_matrix
vars_mean = mean_distr
diff = data - vars_mean
md = []
for i in range(len(diff)):
md.append(np.sqrt(diff[i].dot(inv_covariance_matrix).dot(diff[i])))
return md
def MD_threshold(dist, extreme=False, verbose=False):
# 计算正常运行的马氏距离的阈值,K需要手动确定
k = 3. if extreme else 1.2
threshold = np.mean(dist) * k
return threshold
def get_abnormal_data(dist_train, dist_test, threshold, X_train, X_test):
anomaly_train = pd.DataFrame()
anomaly_train['Mob dist'] = dist_train
anomaly_train['Thresh'] = threshold
# If Mob dist above threshold: Flag as anomaly
anomaly_train['Anomaly'] = anomaly_train['Mob dist'] > anomaly_train['Thresh']
anomaly_train.index = X_train.index
anomaly = pd.DataFrame()
anomaly['Mob dist'] = dist_test
anomaly['Thresh'] = threshold
# If Mob dist above threshold: Flag as anomaly
anomaly['Anomaly'] = anomaly['Mob dist'] > anomaly['Thresh']
anomaly.index = X_test.index
print(anomaly.head())
anomaly_alldata = pd.concat([anomaly_train, anomaly])
return anomaly_alldata
def plot_visible(dist_train):
import seaborn as sns
sns.set(color_codes=True)
# 可视化
plt.figure()
sns.distplot(np.square(dist_train), bins=10, kde=False) # 马氏距离的平方服从卡方分布
plt.xlim([0.0, 15])
plt.xlabel('Mahalanobis dist *2')
plt.show()
plt.figure()
sns.distplot(dist_train, bins=10, kde=True, color='green') # 马氏距离服从正态分布
plt.xlim([0.0, 5])
plt.xlabel('Mahalanobis dist')
plt.show()
if __name__ == '__main__':
param = param
# equipID = param['equipID']
# equipMK = param['equipMK']
# staId = param['staId']
startTime = param['startTime']
endTime = param['endTime']
# iv_data = get_iv_data(startTime, endTime, equipID, staId, equipMK, EQUIP_MK_NAME)
# data = iv_data_process(iv_data, EQUIP_MK_NAME)
data = get_demand_data_finally()
data_original = data.copy()
data = normalize_data(data)
data, columns_name = pca_process_data(data, n_components=n_components)
# # 训练集和测试集
X_train = data.loc[startTime:X_train_end_time]
X_test = data.loc[X_test_start_time:endTime]
cov_matrix, inv_cov_matrix = cov_matrix(X_train.values)
mean_distr = X_train.values.mean(axis=0)
dist_test = MahalanobisDist(inv_cov_matrix, mean_distr, X_test.values, verbose=False)
dist_train = MahalanobisDist(inv_cov_matrix, mean_distr, X_train.values, verbose=False)
# threshold = MD_threshold(dist_train, extreme=True)
threshold = 3.5
plot_visible(dist_train)
anomaly_alldata = get_abnormal_data(dist_train, dist_test, threshold, X_train, X_test)
# 异常数据可视化
anomaly_alldata_finally = anomaly_alldata.copy()
# anomaly_alldata.index = map(lambda x: x[5:10], anomaly_alldata.index)
anomaly_alldata.plot(logy=True, figsize=(10, 6), ylim=[1e-1, 1e3], color=['green', 'red'])
plt.xticks(rotation=0)
plt.show()
anomaly_alldata_finally.to_csv("ma_anomaly_alldata_finally.csv")