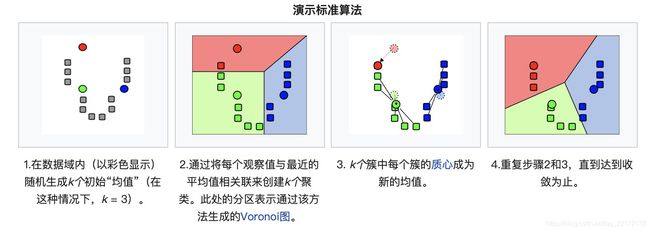
对COVID-19论文进行自动分类——文献聚合分类实现方案
概述
实现步骤:
- 使用自然语言处理(NLP)从每个文档的正文中解析文本。
- 使用术语频率-逆文档频率(TF-IDF)将每个文档实例转换为特征向量 feature。
- 使用 t 分布随机近邻嵌入(t-SNE)对每个特征向量进行降维,将相似的文章聚集在二维平面 1 中。
- 使用主成分分析(PCA)将数据的维数投影到多个维,这些维将保持 0.95 的方差,同时消除嵌入 2 时的噪声和离群值。
- 在 2 上应用 k-means 聚类,其中为 10,以标记 1 上的每个聚类。
- 使用潜在狄利克雷分配(LDA)建模,以从每个聚类中发现关键字。
- 在可视化图形上可视地查找聚类,并使用随机梯度下降(SGD)进行分类。
数据获取和加载
数据集说明:
为了应对COVID-19大流行,白宫和主要研究小组的联盟已经准备好了COVID-19开放研究数据集(CORD-19)。 CORD-19的资源超过300,000篇学术文章,涉及COVID-19,SARS-CoV-2和相关的冠状病毒。 我们本文中采用的就是该数据集。
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import glob
import json
import matplotlib.pyplot as plt
#设置字体、图形样式
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
plt.rcParams['font.sans-serif'] = [u'SimHei']
plt.rcParams['axes.unicode_minus'] = False
plt.rcParams['font.sans-serif'] = ['Arial Unicode MS']
plt.style.use('ggplot')
实验数据
我们可以通过:https://www.kaggle.com/allen-institute-for-ai/CORD-19-research-challenge/download 下载文献数据。下载完成后,先加载文献集的元数据,如下:
root_path = '/Users/shen/Documents/local jupyter/COVID-19/archive/'
metadata_path = f'{root_path}/metadata.csv'
meta_df = pd.read_csv(metadata_path, dtype={
'pubmed_id': str,
'Microsoft Academic Paper ID': str,
'doi': str
})
meta_df.head()
/usr/local/lib/python3.8/site-packages/IPython/core/interactiveshell.py:3145: DtypeWarning: Columns (5,13,14,16) have mixed types.Specify dtype option on import or set low_memory=False.
has_raised = await self.run_ast_nodes(code_ast.body, cell_name,
| cord_uid | sha | source_x | title | doi | pmcid | pubmed_id | license | abstract | publish_time | authors | journal | mag_id | who_covidence_id | arxiv_id | pdf_json_files | pmc_json_files | url | s2_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | ug7v899j | d1aafb70c066a2068b02786f8929fd9c900897fb | PMC | Clinical features of culture-proven Mycoplasma... | 10.1186/1471-2334-1-6 | PMC35282 | 11472636 | no-cc | OBJECTIVE: This retrospective chart review des... | 2001-07-04 | Madani, Tariq A; Al-Ghamdi, Aisha A | BMC Infect Dis | NaN | NaN | NaN | document_parses/pdf_json/d1aafb70c066a2068b027... | document_parses/pmc_json/PMC35282.xml.json | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3... | NaN |
| 1 | 02tnwd4m | 6b0567729c2143a66d737eb0a2f63f2dce2e5a7d | PMC | Nitric oxide: a pro-inflammatory mediator in l... | 10.1186/rr14 | PMC59543 | 11667967 | no-cc | Inflammatory diseases of the respiratory tract... | 2000-08-15 | Vliet, Albert van der; Eiserich, Jason P; Cros... | Respir Res | NaN | NaN | NaN | document_parses/pdf_json/6b0567729c2143a66d737... | document_parses/pmc_json/PMC59543.xml.json | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5... | NaN |
| 2 | ejv2xln0 | 06ced00a5fc04215949aa72528f2eeaae1d58927 | PMC | Surfactant protein-D and pulmonary host defense | 10.1186/rr19 | PMC59549 | 11667972 | no-cc | Surfactant protein-D (SP-D) participates in th... | 2000-08-25 | Crouch, Erika C | Respir Res | NaN | NaN | NaN | document_parses/pdf_json/06ced00a5fc04215949aa... | document_parses/pmc_json/PMC59549.xml.json | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5... | NaN |
| 3 | 2b73a28n | 348055649b6b8cf2b9a376498df9bf41f7123605 | PMC | Role of endothelin-1 in lung disease | 10.1186/rr44 | PMC59574 | 11686871 | no-cc | Endothelin-1 (ET-1) is a 21 amino acid peptide... | 2001-02-22 | Fagan, Karen A; McMurtry, Ivan F; Rodman, David M | Respir Res | NaN | NaN | NaN | document_parses/pdf_json/348055649b6b8cf2b9a37... | document_parses/pmc_json/PMC59574.xml.json | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5... | NaN |
| 4 | 9785vg6d | 5f48792a5fa08bed9f56016f4981ae2ca6031b32 | PMC | Gene expression in epithelial cells in respons... | 10.1186/rr61 | PMC59580 | 11686888 | no-cc | Respiratory syncytial virus (RSV) and pneumoni... | 2001-05-11 | Domachowske, Joseph B; Bonville, Cynthia A; Ro... | Respir Res | NaN | NaN | NaN | document_parses/pdf_json/5f48792a5fa08bed9f560... | document_parses/pmc_json/PMC59580.xml.json | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5... | NaN |
当我们后续将文章聚类,以查看哪些文章聚类在一起时,“title”和“journal”属性可能在以后有用。
meta_df.info()
RangeIndex: 301667 entries, 0 to 301666
Data columns (total 19 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 cord_uid 301667 non-null object
1 sha 111985 non-null object
2 source_x 301667 non-null object
3 title 301587 non-null object
4 doi 186832 non-null object
5 pmcid 116337 non-null object
6 pubmed_id 164429 non-null object
7 license 301667 non-null object
8 abstract 214463 non-null object
9 publish_time 301501 non-null object
10 authors 291797 non-null object
11 journal 282758 non-null object
12 mag_id 0 non-null float64
13 who_covidence_id 99624 non-null object
14 arxiv_id 3884 non-null object
15 pdf_json_files 111985 non-null object
16 pmc_json_files 85059 non-null object
17 url 203159 non-null object
18 s2_id 268534 non-null float64
dtypes: float64(2), object(17)
memory usage: 43.7+ MB
提取文献路径
所有的文献资料已转化为 json 格式的数据,直接加载即可,资料中提供了两种格式的文献资料,分别是PDF的和pmc的,因为数据较多,运行比较耗时,我们这里中选择了PDF的文献。
all_json = glob.glob(f'{root_path}document_parses/pdf_json/*.json', recursive=True)
len(all_json)
118839
一些方法和类的定义
先来定义一个文档阅读器
class FileReader:
def __init__(self, file_path):
with open(file_path) as file:
content = json.load(file)
self.paper_id = content['paper_id']
self.abstract = []
self.body_text = []
# Abstract
for entry in content['abstract']:
self.abstract.append(entry['text'])
# Body text
for entry in content['body_text']:
self.body_text.append(entry['text'])
self.abstract = '\n'.join(self.abstract)
self.body_text = '\n'.join(self.body_text)
def __repr__(self):
return f'{self.paper_id}: {self.abstract[:200]}... {self.body_text[:200]}...'
first_row = FileReader(all_json[0])
print(first_row)
efe13333c69a364cb5d4463ba93815e6fc2d91c6: Background... a1111111111 a1111111111 a1111111111 a1111111111 a1111111111 available data; 78%). In addition, significant increases in the levels of lactate dehydrogenase and α-hydroxybutyrate dehydrogenase were det...
辅助功能会在字符长度达到一定数量时在每个单词后添加断点。
def get_breaks(content, length):
data = ""
words = content.split(' ')
total_chars = 0
# 添加每个长度的字符
for i in range(len(words)):
total_chars += len(words[i])
if total_chars > length:
data = data + "
" + words[i]
total_chars = 0
else:
data = data + " " + words[i]
return data
将数据加载到DataFrame
将文章读入 DataFrame 数据中,这里会比较慢:
%%time
dict_ = {'paper_id': [], 'doi':[], 'abstract': [], 'body_text': [], 'authors': [], 'title': [], 'journal': [], 'abstract_summary': []}
for idx, entry in enumerate(all_json):
if idx % (len(all_json) // 10) == 0:
print(f'Processing index: {idx} of {len(all_json)}')
try:
content = FileReader(entry)
except Exception as e:
continue # 无效的文章格式,跳过
# 获取元数据信息
meta_data = meta_df.loc[meta_df['sha'] == content.paper_id]
# 没有元数据,跳过本文
if len(meta_data) == 0:
continue
dict_['abstract'].append(content.abstract)
dict_['paper_id'].append(content.paper_id)
dict_['body_text'].append(content.body_text)
# 为要在绘图中使用的摘要创建一列
if len(content.abstract) == 0:
# 没有提供摘要
dict_['abstract_summary'].append("Not provided.")
elif len(content.abstract.split(' ')) > 100:
# 提供的摘要太长,取前100个字 + ...
info = content.abstract.split(' ')[:100]
summary = get_breaks(' '.join(info), 40)
dict_['abstract_summary'].append(summary + "...")
else:
summary = get_breaks(content.abstract, 40)
dict_['abstract_summary'].append(summary)
# 获取元数据信息
meta_data = meta_df.loc[meta_df['sha'] == content.paper_id]
try:
# 如果超过一位作者
authors = meta_data['authors'].values[0].split(';')
if len(authors) > 2:
# 如果作者多于2名,在两者之间用html标记分隔
dict_['authors'].append(get_breaks('. '.join(authors), 40))
else:
dict_['authors'].append(". ".join(authors))
except Exception as e:
# 如果只有一位作者或为Null值
dict_['authors'].append(meta_data['authors'].values[0])
# 添加标题信息
try:
title = get_breaks(meta_data['title'].values[0], 40)
dict_['title'].append(title)
# 没有提供标题
except Exception as e:
dict_['title'].append(meta_data['title'].values[0])
# 添加日记信息
dict_['journal'].append(meta_data['journal'].values[0])
# 添加 doi
dict_['doi'].append(meta_data['doi'].values[0])
df_covid = pd.DataFrame(dict_, columns=['paper_id', 'doi', 'abstract', 'body_text', 'authors', 'title', 'journal', 'abstract_summary'])
df_covid.head()
Processing index: 0 of 118839
Processing index: 11883 of 118839
Processing index: 23766 of 118839
Processing index: 35649 of 118839
Processing index: 47532 of 118839
Processing index: 59415 of 118839
Processing index: 71298 of 118839
Processing index: 83181 of 118839
Processing index: 95064 of 118839
Processing index: 106947 of 118839
Processing index: 118830 of 118839
| paper_id | doi | abstract | body_text | authors | title | journal | abstract_summary | |
|---|---|---|---|---|---|---|---|---|
| 0 | efe13333c69a364cb5d4463ba93815e6fc2d91c6 | 10.1371/journal.pmed.1003130 | Background | a1111111111 a1111111111 a1111111111 a111111111... | Zhang, Che. Gu, Jiaowei. Chen, Quanjing. D... | Clinical and epidemiological characteristi... |
PLoS Med | Background |
| 1 | 4fcb95cc0c4ea6d1fa4137a4a087715ed6b68cea | 10.1007/s00431-019-03543-0 | Abnormal levels of end-tidal carbon dioxide (E... | Improvements in neonatal intensive care have r... | Tamura, Kentaro. Williams, Emma E. Dassios,... | End-tidal carbon dioxide levels during res... |
Eur J Pediatr | Abnormal levels of end-tidal carbon dioxide |
| 2 | 94310f437664763acbb472df37158b9694a3bf3a | 10.1371/journal.pone.0236618 | This study aimed to develop risk scores based ... | The coronavirus disease 2019 (COVID- 19) is an... | Zhao, Zirun. Chen, Anne. Hou, Wei. Graham,... | Prediction model and risk scores of ICU ad... |
PLoS One | This study aimed to develop risk scores based... |
| 3 | 86d4262de73cf81b5ea6aafb91630853248bff5f | 10.1016/j.bbamcr.2011.06.011 | The endoplasmic reticulum (ER) is the biggest ... | The endoplasmic reticulum (ER) is a multi-func... | Lynes, Emily M.. Simmen, Thomas | Urban planning of the endoplasmic reticulum| Biochim Biophys Acta Mol Cell Res |
The endoplasmic reticulum (ER) is the biggest... |
|
| 4 | b2f67d533f2749807f2537f3775b39da3b186051 | 10.1016/j.fsiml.2020.100013 | There is a disproportionate number of individu... | Liebrenz, Michael. Bhugra, Dinesh. Buadze,<... | Caring for persons in detention suffering wit... | Forensic Science International: Mind and Law | Not provided. |
熟悉数据
对摘要和正文的字数进行统计
# 摘要中的字数统计
df_covid['abstract_word_count'] = df_covid['abstract'].apply(lambda x: len(x.strip().split()))
# 正文中的字数统计
df_covid['body_word_count'] = df_covid['body_text'].apply(lambda x: len(x.strip().split()))
# 正文中唯一词的统计
df_covid['body_unique_words']=df_covid['body_text'].apply(lambda x:len(set(str(x).split())))
df_covid.head()
| paper_id | doi | abstract | body_text | authors | title | journal | abstract_summary | abstract_word_count | body_word_count | body_unique_words | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | efe13333c69a364cb5d4463ba93815e6fc2d91c6 | 10.1371/journal.pmed.1003130 | Background | a1111111111 a1111111111 a1111111111 a111111111... | Zhang, Che. Gu, Jiaowei. Chen, Quanjing. D... | Clinical and epidemiological characteristi... |
PLoS Med | Background | 1 | 3667 | 1158 |
| 1 | 4fcb95cc0c4ea6d1fa4137a4a087715ed6b68cea | 10.1007/s00431-019-03543-0 | Abnormal levels of end-tidal carbon dioxide (E... | Improvements in neonatal intensive care have r... | Tamura, Kentaro. Williams, Emma E. Dassios,... | End-tidal carbon dioxide levels during res... |
Eur J Pediatr | Abnormal levels of end-tidal carbon dioxide| 218 |
2601 |
830 |
|
| 2 | 94310f437664763acbb472df37158b9694a3bf3a | 10.1371/journal.pone.0236618 | This study aimed to develop risk scores based ... | The coronavirus disease 2019 (COVID- 19) is an... | Zhao, Zirun. Chen, Anne. Hou, Wei. Graham,... | Prediction model and risk scores of ICU ad... |
PLoS One | This study aimed to develop risk scores based... | 225 | 3223 | 1187 |
| 3 | 86d4262de73cf81b5ea6aafb91630853248bff5f | 10.1016/j.bbamcr.2011.06.011 | The endoplasmic reticulum (ER) is the biggest ... | The endoplasmic reticulum (ER) is a multi-func... | Lynes, Emily M.. Simmen, Thomas | Urban planning of the endoplasmic reticulum| Biochim Biophys Acta Mol Cell Res |
The endoplasmic reticulum (ER) is the biggest... |
234 |
8069 |
2282 |
|
| 4 | b2f67d533f2749807f2537f3775b39da3b186051 | 10.1016/j.fsiml.2020.100013 | There is a disproportionate number of individu... | Liebrenz, Michael. Bhugra, Dinesh. Buadze,<... | Caring for persons in detention suffering wit... | Forensic Science International: Mind and Law | Not provided. | 0 | 1126 | 540 |
处理重复项
df_covid.info()
RangeIndex: 105888 entries, 0 to 105887
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 paper_id 105888 non-null object
1 doi 102776 non-null object
2 abstract 105888 non-null object
3 body_text 105888 non-null object
4 authors 104211 non-null object
5 title 105887 non-null object
6 journal 95374 non-null object
7 abstract_summary 105888 non-null object
8 abstract_word_count 105888 non-null int64
9 body_word_count 105888 non-null int64
10 body_unique_words 105888 non-null int64
dtypes: int64(3), object(8)
memory usage: 8.9+ MB
df_covid['abstract'].describe(include='all')
count 105888
unique 71331
top
freq 34018
Name: abstract, dtype: object
根据唯一值,我们可以看到一些重复项,这可能是由于作者将文章提交到多个期刊引起的。我们从数据集中删除重复项:
df_covid.drop_duplicates(['abstract', 'body_text'], inplace=True)
df_covid['abstract'].describe(include='all')
count 105675
unique 71331
top
freq 33877
Name: abstract, dtype: object
df_covid['body_text'].describe(include='all')
count 105675
unique 105668
top J o u r n a l P r e -p r o o f
freq 2
Name: body_text, dtype: object
现在看来没有重复项了。
再来看看数据
df_covid.head()
| paper_id | doi | abstract | body_text | authors | title | journal | abstract_summary | abstract_word_count | body_word_count | body_unique_words | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | efe13333c69a364cb5d4463ba93815e6fc2d91c6 | 10.1371/journal.pmed.1003130 | Background | a1111111111 a1111111111 a1111111111 a111111111... | Zhang, Che. Gu, Jiaowei. Chen, Quanjing. D... | Clinical and epidemiological characteristi... |
PLoS Med | Background | 1 | 3667 | 1158 |
| 1 | 4fcb95cc0c4ea6d1fa4137a4a087715ed6b68cea | 10.1007/s00431-019-03543-0 | Abnormal levels of end-tidal carbon dioxide (E... | Improvements in neonatal intensive care have r... | Tamura, Kentaro. Williams, Emma E. Dassios,... | End-tidal carbon dioxide levels during res... |
Eur J Pediatr | Abnormal levels of end-tidal carbon dioxide| 218 |
2601 |
830 |
|
| 2 | 94310f437664763acbb472df37158b9694a3bf3a | 10.1371/journal.pone.0236618 | This study aimed to develop risk scores based ... | The coronavirus disease 2019 (COVID- 19) is an... | Zhao, Zirun. Chen, Anne. Hou, Wei. Graham,... | Prediction model and risk scores of ICU ad... |
PLoS One | This study aimed to develop risk scores based... | 225 | 3223 | 1187 |
| 3 | 86d4262de73cf81b5ea6aafb91630853248bff5f | 10.1016/j.bbamcr.2011.06.011 | The endoplasmic reticulum (ER) is the biggest ... | The endoplasmic reticulum (ER) is a multi-func... | Lynes, Emily M.. Simmen, Thomas | Urban planning of the endoplasmic reticulum| Biochim Biophys Acta Mol Cell Res |
The endoplasmic reticulum (ER) is the biggest... |
234 |
8069 |
2282 |
|
| 4 | b2f67d533f2749807f2537f3775b39da3b186051 | 10.1016/j.fsiml.2020.100013 | There is a disproportionate number of individu... | Liebrenz, Michael. Bhugra, Dinesh. Buadze,<... | Caring for persons in detention suffering wit... | Forensic Science International: Mind and Law | Not provided. | 0 | 1126 | 540 |
论文主体,使用body_text字段
论文链接,使用doi字段
df_covid.describe()
| abstract_word_count | body_word_count | body_unique_words | |
|---|---|---|---|
| count | 105675.000000 | 105675.000000 | 105675.000000 |
| mean | 153.020251 | 3953.499115 | 1219.518656 |
| std | 196.612981 | 7889.414632 | 1331.522819 |
| min | 0.000000 | 1.000000 | 1.000000 |
| 25% | 0.000000 | 1436.000000 | 641.000000 |
| 50% | 141.000000 | 2872.000000 | 1035.000000 |
| 75% | 231.000000 | 4600.000000 | 1467.000000 |
| max | 7415.000000 | 279623.000000 | 38298.000000 |
数据预处理
现在已经加载了数据集,我们为了聚类效果,需要先清洗文本。首先,删除 Null 值:
df_covid.dropna(inplace=True)
df_covid.info()
Int64Index: 93295 entries, 0 to 105887
Data columns (total 11 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 paper_id 93295 non-null object
1 doi 93295 non-null object
2 abstract 93295 non-null object
3 body_text 93295 non-null object
4 authors 93295 non-null object
5 title 93295 non-null object
6 journal 93295 non-null object
7 abstract_summary 93295 non-null object
8 abstract_word_count 93295 non-null int64
9 body_word_count 93295 non-null int64
10 body_unique_words 93295 non-null int64
dtypes: int64(3), object(8)
memory usage: 8.5+ MB
接下来,我们将确定每篇论文的语言。大部分都是英语,但不全是,因此需要识别语言,以便我们知道如何处理。
from tqdm import tqdm
from langdetect import detect
from langdetect import DetectorFactory
# 设置种子
DetectorFactory.seed = 0
# 保留标签-语言
languages = []
# 文本遍历
for ii in tqdm(range(0,len(df_covid))):
# 按body_text分成列表
text = df_covid.iloc[ii]['body_text'].split(" ")
lang = "en"
try:
if len(text) > 50:
lang = detect(" ".join(text[:50]))
elif len(text) > 0:
lang = detect(" ".join(text[:len(text)]))
# 文档开头的格式不正确
except Exception as e:
all_words = set(text)
try:
lang = detect(" ".join(all_words))
except Exception as e:
try:
# 通过摘要标记
lang = detect(df_covid.iloc[ii]['abstract_summary'])
except Exception as e:
lang = "unknown"
pass
languages.append(lang)
100%|██████████| 93295/93295 [09:31<00:00, 163.21it/s]
from pprint import pprint
languages_dict = {}
for lang in set(languages):
languages_dict[lang] = languages.count(lang)
print("Total: {}\n".format(len(languages)))
pprint(languages_dict)
Total: 93295
{'af': 3,
'ca': 4,
'cy': 7,
'de': 1158,
'en': 90495,
'es': 840,
'et': 1,
'fr': 566,
'id': 1,
'it': 42,
'nl': 123,
'pl': 1,
'pt': 43,
'ro': 1,
'sq': 1,
'sv': 1,
'tl': 2,
'tr': 1,
'vi': 1,
'zh-cn': 4}
看一下各语言的分布情况:
df_covid['language'] = languages
plt.bar(range(len(languages_dict)), list(languages_dict.values()), align='center')
plt.xticks(range(len(languages_dict)), list(languages_dict.keys()))
plt.title("Distribution of Languages in Dataset")
plt.show()
其他语言的论文很少,删除所有非英语的语言:
df_covid = df_covid[df_covid['language'] == 'en']
df_covid.info()
Int64Index: 90495 entries, 0 to 105887
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 paper_id 90495 non-null object
1 doi 90495 non-null object
2 abstract 90495 non-null object
3 body_text 90495 non-null object
4 authors 90495 non-null object
5 title 90495 non-null object
6 journal 90495 non-null object
7 abstract_summary 90495 non-null object
8 abstract_word_count 90495 non-null int64
9 body_word_count 90495 non-null int64
10 body_unique_words 90495 non-null int64
11 language 90495 non-null object
dtypes: int64(3), object(9)
memory usage: 9.0+ MB
# 下载 spacy 解析器
from IPython.utils import io
with io.capture_output() as captured:
!pip install https://s3-us-west-2.amazonaws.com/ai2-s2-scispacy/releases/v0.2.4/en_core_sci_lg-0.2.4.tar.gz
#NLP
import spacy
from spacy.lang.en.stop_words import STOP_WORDS
import en_core_sci_lg
/usr/local/lib/python3.8/site-packages/spacy/util.py:275: UserWarning: [W031] Model 'en_core_sci_lg' (0.2.4) requires spaCy v2.2 and is incompatible with the current spaCy version (2.3.2). This may lead to unexpected results or runtime errors. To resolve this, download a newer compatible model or retrain your custom model with the current spaCy version. For more details and available updates, run: python -m spacy validate
warnings.warn(warn_msg)
为了在聚类时降噪,删除英文停用词(一些没有实际含义的词,比如the a it等)
import string
punctuations = string.punctuation
stopwords = list(STOP_WORDS)
stopwords[:10]
['’s',
'sometimes',
'until',
'may',
'most',
'each',
'formerly',
'nevertheless',
'ever',
'though']
custom_stop_words = [
'doi', 'preprint', 'copyright', 'peer', 'reviewed', 'org', 'https', 'et', 'al', 'author', 'figure',
'rights', 'reserved', 'permission', 'used', 'using', 'biorxiv', 'medrxiv', 'license', 'fig', 'fig.',
'al.', 'Elsevier', 'PMC', 'CZI', 'www'
]
for w in custom_stop_words:
if w not in stopwords:
stopwords.append(w)
接下来,创建一个处理文本数据的函数。该函数会将文本转换为小写字母,删除标点符号和停用词。对于解析器,使用en_core_sci_lg。
parser = en_core_sci_lg.load(disable=["tagger", "ner"])
parser.max_length = 7000000
def spacy_tokenizer(sentence):
mytokens = parser(sentence)
mytokens = [ word.lemma_.lower().strip() if word.lemma_ != "-PRON-" else word.lower_ for word in mytokens ]
mytokens = [ word for word in mytokens if word not in stopwords and word not in punctuations ]
mytokens = " ".join([i for i in mytokens])
return mytokens
在body_text上应用文本处理功能。
tqdm.pandas()
df_covid["processed_text"] = df_covid["body_text"].progress_apply(spacy_tokenizer)
/usr/local/lib/python3.8/site-packages/tqdm/std.py:697: FutureWarning: The Panel class is removed from pandas. Accessing it from the top-level namespace will also be removed in the next version
from pandas import Panel
100%|██████████| 90495/90495 [6:34:56<00:00, 3.82it/s]
让我们看一下论文中的字数统计:
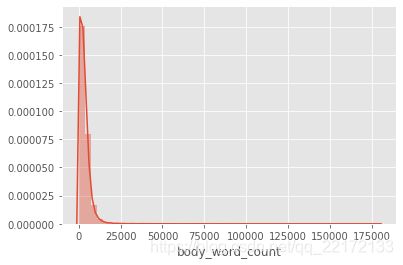
import seaborn as sns
sns.distplot(df_covid['body_word_count'])
df_covid['body_word_count'].describe()
count 90495.000000
mean 3529.074413
std 3817.917262
min 1.000000
25% 1402.500000
50% 2851.000000
75% 4550.000000
max 179548.000000
Name: body_word_count, dtype: float64
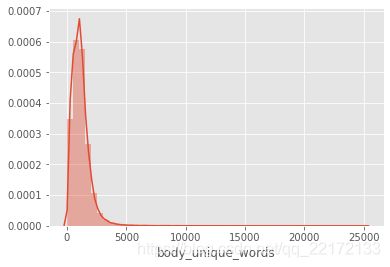
sns.distplot(df_covid['body_unique_words'])
df_covid['body_unique_words'].describe()
count 90495.000000
mean 1153.912758
std 813.610719
min 1.000000
25% 635.000000
50% 1037.000000
75% 1465.000000
max 25156.000000
Name: body_unique_words, dtype: float64
这两幅图使我们对正在处理的内容有了一个很好的了解。 大多数论文的长度约为5000字。 两个图中的长尾巴都是由异常值引起的。 实际上,约有98%的论文篇幅少于20,000个字,而少数论文则超过200,000个!
向量化
目前,我们已经对数据进行了预处理,现在将其转换为可以由我们算法处理的格式。
我们将使用TF/IDF,这是衡量每个单词对整个文献重要性衡量的标准方式。
from sklearn.feature_extraction.text import TfidfVectorizer
def vectorize(text, maxx_features):
vectorizer = TfidfVectorizer(max_features=maxx_features)
X = vectorizer.fit_transform(text)
return X
向量化我们的数据,我们将基于正文的内容进行聚类。特征最大数量我们限制为前2 ** 12,首先为了降低噪声,此外,太多的话运行时间也会过长。
text = df_covid['processed_text'].values
X = vectorize(text, 2 ** 12)
X.shape
(90495, 4096)
PCA
对矢量数据进行主成分分析(PCA)。这样做的原因是,通过PCA保留大部分信息,但是可以从数据中消除一些噪音/离群值,使k-means的聚类更加容易。
注意,X_reduced仅用于k-均值,t-SNE仍使用通过tf-idf对NLP处理的文本生成的原始特征向量X。
from sklearn.decomposition import PCA
pca = PCA(n_components=0.95, random_state=42)
X_reduced= pca.fit_transform(X.toarray())
X_reduced.shape
(90495, 2793)
聚类划分
为了分开文献,将在矢量文本上运行k-means。 给定簇数k,k-means将通过取平均距离到随机初始化的质心的方式对每个向量进行分类,重心迭代更新。
from sklearn.cluster import KMeans
我们需要找到最佳k值,具体做法是让k从1开始取值直到取到你认为合适的上限(一般来说这个上限不会太大,这里因为文献类目较多,我们选取上限为50),对每一个k值进行聚类并且记下对于的SSE,然后画出k和SSE的关系图(手肘形),最后选取肘部对应的k作为我们的最佳聚类数。
from sklearn import metrics
from scipy.spatial.distance import cdist
# 用不同的k运行kmeans
distortions = []
K = range(2, 50)
for k in K:
k_means = KMeans(n_clusters=k, random_state=42).fit(X_reduced)
k_means.fit(X_reduced)
distortions.append(sum(np.min(cdist(X_reduced, k_means.cluster_centers_, 'euclidean'), axis=1)) / X.shape[0])
X_line = [K[0], K[-1]]
Y_line = [distortions[0], distortions[-1]]
plt.plot(K, distortions, 'b-')
plt.plot(X_line, Y_line, 'r')
plt.xlabel('k')
plt.ylabel('Distortion')
plt.title('肘法显示最优k')
plt.show()
在该图中,我们可以看到较好的k值在8-15之间。 此后,失真的降低就不那么明显了。 为简单起见,我们将使用k = 10。现在我们有了一个合适的k值,然后在经过PCA处理的特征向量(X_reduced)上运行k-means:
k = 10
kmeans = KMeans(n_clusters=k, random_state=42)
y_pred = kmeans.fit_predict(X_reduced)
df_covid['y'] = y_pred
使用t-SNE降维
t分布随机邻域嵌入(t-distributed stochastic neighbor embedding,t-SNE),是一种用于探索高维数据的非线性降维机器学习算法。它将多维数据映射到适合于人类观察的两个或多个维度。PCA是一种线性算法,它不能解释特征之间的复杂多项式关系。而t-SNE是基于在邻域图上随机游走的概率分布来找到数据内的结构。
from sklearn.manifold import TSNE
tsne = TSNE(verbose=1, perplexity=100, random_state=42)
X_embedded = tsne.fit_transform(X.toarray())
[t-SNE] Computing 301 nearest neighbors...
[t-SNE] Indexed 90495 samples in 114.698s...
[t-SNE] Computed neighbors for 90495 samples in 51904.808s...
[t-SNE] Computed conditional probabilities for sample 1000 / 90495
[t-SNE] Computed conditional probabilities for sample 2000 / 90495
[t-SNE] Computed conditional probabilities for sample 3000 / 90495
[t-SNE] Computed conditional probabilities for sample 4000 / 90495
[t-SNE] Computed conditional probabilities for sample 5000 / 90495
[t-SNE] Computed conditional probabilities for sample 6000 / 90495
[t-SNE] Computed conditional probabilities for sample 7000 / 90495
[t-SNE] Computed conditional probabilities for sample 8000 / 90495
[t-SNE] Computed conditional probabilities for sample 9000 / 90495
[t-SNE] Computed conditional probabilities for sample 10000 / 90495
[t-SNE] Computed conditional probabilities for sample 11000 / 90495
[t-SNE] Computed conditional probabilities for sample 12000 / 90495
[t-SNE] Computed conditional probabilities for sample 13000 / 90495
[t-SNE] Computed conditional probabilities for sample 14000 / 90495
[t-SNE] Computed conditional probabilities for sample 15000 / 90495
[t-SNE] Computed conditional probabilities for sample 16000 / 90495
[t-SNE] Computed conditional probabilities for sample 17000 / 90495
[t-SNE] Computed conditional probabilities for sample 18000 / 90495
[t-SNE] Computed conditional probabilities for sample 19000 / 90495
[t-SNE] Computed conditional probabilities for sample 20000 / 90495
[t-SNE] Computed conditional probabilities for sample 21000 / 90495
[t-SNE] Computed conditional probabilities for sample 22000 / 90495
[t-SNE] Computed conditional probabilities for sample 23000 / 90495
[t-SNE] Computed conditional probabilities for sample 24000 / 90495
[t-SNE] Computed conditional probabilities for sample 25000 / 90495
[t-SNE] Computed conditional probabilities for sample 26000 / 90495
[t-SNE] Computed conditional probabilities for sample 27000 / 90495
[t-SNE] Computed conditional probabilities for sample 28000 / 90495
[t-SNE] Computed conditional probabilities for sample 29000 / 90495
[t-SNE] Computed conditional probabilities for sample 30000 / 90495
[t-SNE] Computed conditional probabilities for sample 31000 / 90495
[t-SNE] Computed conditional probabilities for sample 32000 / 90495
[t-SNE] Computed conditional probabilities for sample 33000 / 90495
[t-SNE] Computed conditional probabilities for sample 34000 / 90495
[t-SNE] Computed conditional probabilities for sample 35000 / 90495
[t-SNE] Computed conditional probabilities for sample 36000 / 90495
[t-SNE] Computed conditional probabilities for sample 37000 / 90495
[t-SNE] Computed conditional probabilities for sample 38000 / 90495
[t-SNE] Computed conditional probabilities for sample 39000 / 90495
[t-SNE] Computed conditional probabilities for sample 40000 / 90495
[t-SNE] Computed conditional probabilities for sample 41000 / 90495
[t-SNE] Computed conditional probabilities for sample 42000 / 90495
[t-SNE] Computed conditional probabilities for sample 43000 / 90495
[t-SNE] Computed conditional probabilities for sample 44000 / 90495
[t-SNE] Computed conditional probabilities for sample 45000 / 90495
[t-SNE] Computed conditional probabilities for sample 46000 / 90495
[t-SNE] Computed conditional probabilities for sample 47000 / 90495
[t-SNE] Computed conditional probabilities for sample 48000 / 90495
[t-SNE] Computed conditional probabilities for sample 49000 / 90495
[t-SNE] Computed conditional probabilities for sample 50000 / 90495
[t-SNE] Computed conditional probabilities for sample 51000 / 90495
[t-SNE] Computed conditional probabilities for sample 52000 / 90495
[t-SNE] Computed conditional probabilities for sample 53000 / 90495
[t-SNE] Computed conditional probabilities for sample 54000 / 90495
[t-SNE] Computed conditional probabilities for sample 55000 / 90495
[t-SNE] Computed conditional probabilities for sample 56000 / 90495
[t-SNE] Computed conditional probabilities for sample 57000 / 90495
[t-SNE] Computed conditional probabilities for sample 58000 / 90495
[t-SNE] Computed conditional probabilities for sample 59000 / 90495
[t-SNE] Computed conditional probabilities for sample 60000 / 90495
[t-SNE] Computed conditional probabilities for sample 61000 / 90495
[t-SNE] Computed conditional probabilities for sample 62000 / 90495
[t-SNE] Computed conditional probabilities for sample 63000 / 90495
[t-SNE] Computed conditional probabilities for sample 64000 / 90495
[t-SNE] Computed conditional probabilities for sample 65000 / 90495
[t-SNE] Computed conditional probabilities for sample 66000 / 90495
[t-SNE] Computed conditional probabilities for sample 67000 / 90495
[t-SNE] Computed conditional probabilities for sample 68000 / 90495
[t-SNE] Computed conditional probabilities for sample 69000 / 90495
[t-SNE] Computed conditional probabilities for sample 70000 / 90495
[t-SNE] Computed conditional probabilities for sample 71000 / 90495
[t-SNE] Computed conditional probabilities for sample 72000 / 90495
[t-SNE] Computed conditional probabilities for sample 73000 / 90495
[t-SNE] Computed conditional probabilities for sample 74000 / 90495
[t-SNE] Computed conditional probabilities for sample 75000 / 90495
[t-SNE] Computed conditional probabilities for sample 76000 / 90495
[t-SNE] Computed conditional probabilities for sample 77000 / 90495
[t-SNE] Computed conditional probabilities for sample 78000 / 90495
[t-SNE] Computed conditional probabilities for sample 79000 / 90495
[t-SNE] Computed conditional probabilities for sample 80000 / 90495
[t-SNE] Computed conditional probabilities for sample 81000 / 90495
[t-SNE] Computed conditional probabilities for sample 82000 / 90495
[t-SNE] Computed conditional probabilities for sample 83000 / 90495
[t-SNE] Computed conditional probabilities for sample 84000 / 90495
[t-SNE] Computed conditional probabilities for sample 85000 / 90495
[t-SNE] Computed conditional probabilities for sample 86000 / 90495
[t-SNE] Computed conditional probabilities for sample 87000 / 90495
[t-SNE] Computed conditional probabilities for sample 88000 / 90495
[t-SNE] Computed conditional probabilities for sample 89000 / 90495
[t-SNE] Computed conditional probabilities for sample 90000 / 90495
[t-SNE] Computed conditional probabilities for sample 90495 / 90495
[t-SNE] Mean sigma: 0.324191
[t-SNE] KL divergence after 250 iterations with early exaggeration: 110.000687
[t-SNE] KL divergence after 1000 iterations: 3.334843
让我们看一下数据压缩为2维后的样子:
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
plt.rcParams['font.family'] = ['Arial Unicode MS'] #用来正常显示中文标签
plt.rcParams['axes.unicode_minus'] = False #用来正常显示负号
sns.set_style('whitegrid',{'font.sans-serif':['Arial Unicode MS','Arial']})
# 设置颜色
palette = sns.color_palette("bright", 1)
plt.figure(figsize=(15, 15))
sns.scatterplot(X_embedded[:,0], X_embedded[:,1], palette=palette)
plt.title('无标签显示 t-SNE')
plt.savefig("./t-sne_covid19.png")
plt.show()
从图上也看不出什么来,我们可以看到一些簇,但是靠近中心的许多实例很难分离。t-SNE在降低尺寸方面做得很好,但是现在我们需要一些标签。我们可以使用k-means发现的聚类作为标签,这样有助于从视觉上区分文献主题。
# 设置颜色
palette = sns.hls_palette(10, l=.4, s=.9)
plt.figure(figsize=(15, 15))
sns.scatterplot(X_embedded[:,0], X_embedded[:,1], hue=y_pred, legend='full', palette=palette)
plt.title('带有Kmeans标签的t-SNE')
plt.savefig("./improved_cluster_tsne.png")
plt.show()
这张图就能很好的区分文献的分组方式,即使k-means和t-SNE是独立运行的,但是它们也能够在集群上达成共识。通过t-SNE可以确定每个文献在图中的位置,而通过k-means则可以确定标签(颜色)。不过,也有一些标签(k-means)很分散的散布在绘图上(t-SNE),因为有些文献的主题经常相交,很难将它们清晰地分开。
作为一种无监督的方法,这些算法可以找到人类不熟悉的数据划分方式。通过对划分结果的研究,我们可能会发现一些隐藏的数据信息,从而推进进一步的研究。当然数据的这种组织方式并不能充当简单的搜索引擎,我们目前只是对文献的数学相似性执行聚类和降维。
主题建模
现在,我们将尝试在每个聚类中找到可以充当关键字的词。K-means将文章聚类,但未标记主题。通过主题建模,我们将发现每个集群最重要的关键字是什么。通过提供关键字以快速识别集群的主题。
对于主题建模,我们将使用LDA(隐含狄利克雷分布)。 在LDA中,每个文档都可以通过主题分布来描述,每个主题都可以通过单词分布来描述。首先,我们将创建10个矢量化容器,每个集群标签一个。
from sklearn.decomposition import LatentDirichletAllocation
from sklearn.feature_extraction.text import CountVectorizer
vectorizers = []
for ii in range(0, 10):
# Creating a vectorizer
vectorizers.append(CountVectorizer(min_df=5, max_df=0.9, stop_words='english', lowercase=True, token_pattern='[a-zA-Z\-][a-zA-Z\-]{2,}'))
vectorizers[0]
CountVectorizer(max_df=0.9, min_df=5, stop_words='english',
token_pattern='[a-zA-Z\\-][a-zA-Z\\-]{2,}')
现在,我们将对每个集群的数据进行矢量化处理:
vectorized_data = []
for current_cluster, cvec in enumerate(vectorizers):
try:
vectorized_data.append(cvec.fit_transform(df_covid.loc[df_covid['y'] == current_cluster, 'processed_text']))
except Exception as e:
print("集群中实例不足: " + str(current_cluster))
vectorized_data.append(None)
len(vectorized_data)
10
主题建模使用LDA来完成,可以通过共享的主题来解释集群。
NUM_TOPICS_PER_CLUSTER = 10
lda_models = []
for ii in range(0, NUM_TOPICS_PER_CLUSTER):
# 隐含狄利克雷分布模型
lda = LatentDirichletAllocation(n_components=NUM_TOPICS_PER_CLUSTER, max_iter=10, learning_method='online',verbose=False, random_state=42)
lda_models.append(lda)
lda_models[0]
LatentDirichletAllocation(learning_method='online', random_state=42,
verbose=False)
对于每个集群,我们在上一步中创建了一个对应的LDA模型。现在,我们将对所有LDA模型进行适当的聚类转换。
clusters_lda_data = []
for current_cluster, lda in enumerate(lda_models):
if vectorized_data[current_cluster] != None:
clusters_lda_data.append((lda.fit_transform(vectorized_data[current_cluster])))
从每个群集中提取关键字:
# 输出每个主题的关键字的功能:
def selected_topics(model, vectorizer, top_n=3):
current_words = []
keywords = []
for idx, topic in enumerate(model.components_):
words = [(vectorizer.get_feature_names()[i], topic[i]) for i in topic.argsort()[:-top_n - 1:-1]]
for word in words:
if word[0] not in current_words:
keywords.append(word)
current_words.append(word[0])
keywords.sort(key = lambda x: x[1])
keywords.reverse()
return_values = []
for ii in keywords:
return_values.append(ii[0])
return return_values
将单个群集的关键字列表追加到长度为NUM_TOPICS_PER_CLUSTER的列表中:
all_keywords = []
for current_vectorizer, lda in enumerate(lda_models):
if vectorized_data[current_vectorizer] != None:
all_keywords.append(selected_topics(lda, vectorizers[current_vectorizer]))
all_keywords[0][:10]
['ang',
'covid-',
'patient',
'bind',
'protein',
'lung',
'sars-cov-',
'gene',
'tmprss',
'effect']
len(all_keywords)
10
我们先把当前输出内容保存到文件,不然重新运行上面的内容非常耗时(我断断续续的运行了两三天,尤其是矢量化和t-SNE)。
f=open('topics.txt','w')
count = 0
for ii in all_keywords:
if vectorized_data[count] != None:
f.write(', '.join(ii) + "\n")
else:
f.write("实例数量不足。 \n")
f.write(', '.join(ii) + "\n")
count += 1
f.close()
import pickle
# 保存COVID-19 DataFrame
pickle.dump(df_covid, open("df_covid.p", "wb" ))
# 保存最终的t-SNE
pickle.dump(X_embedded, open("X_embedded.p", "wb" ))
# 保存用k-means生成的标签(10)
pickle.dump(y_pred, open("y_pred.p", "wb" ))
分类
在运行kmeans之后,现在已对数据进行“标记”。这意味着我们现在可以使用监督学习来了解聚类的概括程度。这只是评估聚类的一种方法,如果k-means能够在数据中找到有意义的拆分,则应该可以训练分类器来预测给定实例应属于哪个聚类。
# 打印分类模型报告
def classification_report(model_name, test, pred):
from sklearn.metrics import precision_score, recall_score
from sklearn.metrics import accuracy_score
from sklearn.metrics import f1_score
print(model_name, ":\n")
print("Accuracy Score: ", '{:,.3f}'.format(float(accuracy_score(test, pred)) * 100), "%")
print(" Precision: ", '{:,.3f}'.format(float(precision_score(test, pred, average='macro')) * 100), "%")
print(" Recall: ", '{:,.3f}'.format(float(recall_score(test, pred, average='macro')) * 100), "%")
print(" F1 score: ", '{:,.3f}'.format(float(f1_score(test, pred, average='macro')) * 100), "%")
划分训练集和测试集
from sklearn.model_selection import train_test_split
# 测试集大小为数据的20%,随机种子为42
X_train, X_test, y_train, y_test = train_test_split(X.toarray(),y_pred, test_size=0.2, random_state=42)
print("X_train size:", len(X_train))
print("X_test size:", len(X_test), "\n")
X_train size: 72396
X_test size: 18099
精确率:预测结果为正例样本中真实为正例的比例(查得准);
召回率:真实为正例的样本中预测结果为正例的比例(查的全,对正样本的区分能力);
F1分数:精度和查全率的谐波平均值,只有精度和召回率都很高时,F1分数才会很高。
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import cross_val_predict
from sklearn.linear_model import SGDClassifier
# SGD 实例
sgd_clf = SGDClassifier(max_iter=10000, tol=1e-3, random_state=42, n_jobs=4)
# 训练 SGD
sgd_clf.fit(X_train, y_train)
# 交叉验证
sgd_pred = cross_val_predict(sgd_clf, X_train, y_train, cv=3, n_jobs=4)
# 分类报告
classification_report("随机梯度下降报告(训练集)", y_train, sgd_pred)
随机梯度下降报告(训练集) :
Accuracy Score: 93.015 %
Precision: 93.202 %
Recall: 92.543 %
F1 score: 92.829 %
然后看看测试集表现如何:
sgd_pred = cross_val_predict(sgd_clf, X_test, y_test, cv=3, n_jobs=4)
classification_report("随机梯度下降报告(测试集)", y_test, sgd_pred)
随机梯度下降报告(测试集) :
Accuracy Score: 91.292 %
Precision: 91.038 %
Recall: 90.799 %
F1 score: 90.887 %
现在,让我们看看模型在整个数据集中如何:
sgd_cv_score = cross_val_score(sgd_clf, X.toarray(), y_pred, cv=10)
print("Mean cv Score - SGD: {:,.3f}".format(float(sgd_cv_score.mean()) * 100), "%")
Mean cv Score - SGD: 93.484 %
BokehJS 绘制数据
前面的步骤为我们提供了聚类标签和二维的论文数据集。然后通过k-means,我们可以看到论文的划分情况。为了理解同一聚类的相似之处,我们还对每组论文进行了主题建模,以挑选出关键词。
现在我们会使用Bokeh进行绘图,它会将实际论文与其在t-SNE图上的位置配对。通过这种方法,将更容易看到这些论文是如何组合在一起的,从而可以研究数据集和评估聚类。
import os
# 切换到lib目录以加载绘图python脚本
main_path = os.getcwd()
lib_path = '/Users/shen/Documents/local jupyter/COVID-19/archive/resources'
os.chdir(lib_path)
# 绘图所需的库
import bokeh
from bokeh.models import ColumnDataSource, HoverTool, LinearColorMapper, CustomJS, Slider, TapTool, TextInput
from bokeh.palettes import Category20
from bokeh.transform import linear_cmap, transform
from bokeh.io import output_file, show, output_notebook
from bokeh.plotting import figure
from bokeh.models import RadioButtonGroup, TextInput, Div, Paragraph
from bokeh.layouts import column, widgetbox, row, layout
from bokeh.layouts import column
os.chdir(main_path)
函数加载:
# 处理当前选择的文章
def selected_code():
code = """
var titles = [];
var authors = [];
var journals = [];
var links = [];
cb_data.source.selected.indices.forEach(index => titles.push(source.data['titles'][index]));
cb_data.source.selected.indices.forEach(index => authors.push(source.data['authors'][index]));
cb_data.source.selected.indices.forEach(index => journals.push(source.data['journal'][index]));
cb_data.source.selected.indices.forEach(index => links.push(source.data['links'][index]));
var title = "" + titles[0].toString().replace(/
/g, ' ') + "
";
var authors = "作者: " + authors[0].toString().replace(/
/g, ' ') + "
"
// var journal = "刊物:" + journals[0].toString() + "
"
var link = "文章链接: " + "http://doi.org/" + links[0].toString() + " "
current_selection.text = title + authors + link
current_selection.change.emit();
"""
return code
# 处理关键字并搜索
def input_callback(plot, source, out_text, topics):
# slider call back
callback = CustomJS(args=dict(p=plot, source=source, out_text=out_text, topics=topics), code="""
var key = text.value;
key = key.toLowerCase();
var cluster = slider.value;
var data = source.data;
console.log(cluster);
var x = data['x'];
var y = data['y'];
var x_backup = data['x_backup'];
var y_backup = data['y_backup'];
var labels = data['desc'];
var abstract = data['abstract'];
var titles = data['titles'];
var authors = data['authors'];
var journal = data['journal'];
if (cluster == '10') {
out_text.text = '关键词:滑动到特定的群集以查看关键词。';
for (var i = 0; i < x.length; i++) {
if(abstract[i].includes(key) ||
titles[i].includes(key) ||
authors[i].includes(key) ||
journal[i].includes(key)) {
x[i] = x_backup[i];
y[i] = y_backup[i];
} else {
x[i] = undefined;
y[i] = undefined;
}
}
}
else {
out_text.text = '关键词: ' + topics[Number(cluster)];
for (var i = 0; i < x.length; i++) {
if(labels[i] == cluster) {
if(abstract[i].includes(key) ||
titles[i].includes(key) ||
authors[i].includes(key) ||
journal[i].includes(key)) {
x[i] = x_backup[i];
y[i] = y_backup[i];
} else {
x[i] = undefined;
y[i] = undefined;
}
} else {
x[i] = undefined;
y[i] = undefined;
}
}
}
source.change.emit();
""")
return callback
每个集群加载关键字:
import os
topic_path = 'topics.txt'
with open(topic_path) as f:
topics = f.readlines()
# 在notebook中显示
output_notebook()
# 目标标签
y_labels = y_pred
# 数据源
source = ColumnDataSource(data=dict(
x= X_embedded[:,0],
y= X_embedded[:,1],
x_backup = X_embedded[:,0],
y_backup = X_embedded[:,1],
desc= y_labels,
titles= df_covid['title'],
authors = df_covid['authors'],
journal = df_covid['journal'],
abstract = df_covid['abstract_summary'],
labels = ["C-" + str(x) for x in y_labels],
links = df_covid['doi']
))
# 鼠标悬停的信息显示
hover = HoverTool(tooltips=[
("标题", "@titles{safe}"),
("作者", "@authors{safe}"),
("出版物", "@journal"),
("简介", "@abstract{safe}"),
("文章链接", "@links")
],
point_policy="follow_mouse")
# 绘图颜色
mapper = linear_cmap(field_name='desc',
palette=Category20[10],
low=min(y_labels) ,high=max(y_labels))
# 大小
plot = figure(plot_width=1200, plot_height=850,
tools=[hover, 'pan', 'wheel_zoom', 'box_zoom', 'reset', 'save', 'tap'],
title="用t-SNE和K-Means对COVID-19文献进行聚类",
toolbar_location="above")
# 绘图
plot.scatter('x', 'y', size=5,
source=source,
fill_color=mapper,
line_alpha=0.3,
line_color="black",
legend = 'labels')
plot.legend.background_fill_alpha = 0.6
BokehDeprecationWarning: 'legend' keyword is deprecated, use explicit 'legend_label', 'legend_field', or 'legend_group' keywords instead
部件加载:
# 关键字
text_banner = Paragraph(text= '关键词:滑动到特定的群集以查看关键词。', height=45)
input_callback_1 = input_callback(plot, source, text_banner, topics)
# 当前选择的文章
div_curr = Div(text="""单击图解以查看文章的链接。""",height=150)
callback_selected = CustomJS(args=dict(source=source, current_selection=div_curr), code=selected_code())
taptool = plot.select(type=TapTool)
taptool.callback = callback_selected
# 工具栏
# slider = Slider(start=0, end=10, value=10, step=1, title="簇 #", callback=input_callback_1)
slider = Slider(start=0, end=10, value=10, step=1, title="簇 #")
# slider.callback = input_callback_1
keyword = TextInput(title="搜索:")
# keyword.callback = input_callback_1
# 回掉参数
input_callback_1.args["text"] = keyword
input_callback_1.args["slider"] = slider
slider.js_on_change('value', input_callback_1)
keyword.js_on_change('value', input_callback_1)
样式设置:
slider.sizing_mode = "stretch_width"
slider.margin=15
keyword.sizing_mode = "scale_both"
keyword.margin=15
div_curr.style={'color': '#BF0A30', 'font-family': 'Helvetica Neue, Helvetica, Arial, sans-serif;', 'font-size': '1.1em'}
div_curr.sizing_mode = "scale_both"
div_curr.margin = 20
text_banner.style={'color': '#0269A4', 'font-family': 'Helvetica Neue, Helvetica, Arial, sans-serif;', 'font-size': '1.1em'}
text_banner.sizing_mode = "scale_both"
text_banner.margin = 20
plot.sizing_mode = "scale_both"
plot.margin = 5
r = row(div_curr,text_banner)
r.sizing_mode = "stretch_width"
显示:
# 页面布局
l = layout([
[slider, keyword],
# [slider],
[text_banner],
[div_curr],
[plot],
])
l.sizing_mode = "scale_both"
output_file('t-sne_covid-19_interactive.html')
show(l)
这里没法放html片段,所以我把最终结果制作成动图展示: