文章目录
- 提醒
- 代码
-
- 数据处理
- 分离 data 和 label
- 训练
- 训练结果 & 混淆矩阵
- 各种 feature 的重要性
提醒
- pandas 读取 excel 文件,需要
xlrd >= 1.1.0
代码
import sklearn
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from sklearn.model_selection import cross_val_score
from sklearn.metrics import roc_curve, f1_score, precision_score, recall_score
from sklearn.svm import SVC
pd.set_option('display.max_rows', None)
pd.set_option('display.max_columns', None)
pd.set_option('display.width', None)
pd.set_option('display.max_colwidth', -1)
数据处理
path = "./2019-2020年.xlsx"
df = pd.read_excel(path)
df = df.drop(['OTT', 'TOAST subtypes'],axis=1)
df = df.dropna(axis=0, how='any')
df = df.reset_index()
df['Coronary heart disease'][74] = 0
df.head(10)
|
index |
Sex |
Medication before thrombolytic therapy |
Age |
Age.1 |
Periventricular White Matter |
Deep White Matter |
The degree of WMH |
Smoking |
Drinking |
AtrialFibrillation |
Hypertension |
Diabetes |
Hyperlipidemia |
Coronary heart disease |
Heart failure |
Stroke |
TIA |
WBC |
N |
L |
NLR |
HB |
PLT |
PCV |
PT |
INR |
APTT |
TT |
Fibrinogen |
Emergency blood sugar |
Fasting blood glucose |
Creatinine |
HDL |
LDL |
HDL/LDL比值 |
Cholesterol |
Triglyceride |
HBLAC |
HCY |
DNT |
sBP |
dBP |
Baseline NIHSS score |
Hemorrhagic Transformation(HT) |
Early neurological deterioration (END) |
Prognosis&0 (mRS0-2:0;3-6:1) |
90dmRS |
Prognosis&1(mRS0-1:0;2-6:1) |
| 0 |
5 |
0 |
2 |
58.0 |
0.0 |
2 |
1 |
2 |
1.0 |
1.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
8.4 |
5.44 |
2.35 |
2.310000 |
141 |
266 |
38 |
10.7 |
0.92 |
34.5 |
14.9 |
3.52 |
5.68 |
5.11 |
66.2 |
0.92 |
3.15 |
0.290000 |
4.27 |
1.99 |
4.6 |
11.0 |
25.0 |
177.0 |
90.0 |
8 |
0 |
0 |
1 |
5 |
1 |
| 1 |
6 |
0 |
2 |
53.0 |
0.0 |
2 |
1 |
2 |
1.0 |
1.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
11.6 |
6.5 |
4.04 |
1.608911 |
145 |
259 |
45 |
9.9 |
0.86 |
30.6 |
15.3 |
3.34 |
6.38 |
6.13 |
74.0 |
1.46 |
4.54 |
0.321586 |
6.11 |
2.39 |
5.5 |
11.3 |
37.0 |
166.0 |
98.0 |
4 |
0 |
0 |
0 |
1 |
0 |
| 2 |
10 |
1 |
2 |
77.0 |
1.0 |
3 |
3 |
3 |
0.0 |
0.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
1 |
0.0 |
7.83 |
6.63 |
0.59 |
11.237288 |
103 |
225 |
30 |
9.2 |
0.80 |
28 |
14.9 |
4.82 |
1.76 |
7.11 |
107.9 |
1.19 |
1.03 |
1.155340 |
2.28 |
0.41 |
5.0 |
13.0 |
30.0 |
150.0 |
90.0 |
9 |
0 |
0 |
0 |
0 |
0 |
| 3 |
13 |
0 |
2 |
65.0 |
0.0 |
1 |
0 |
1 |
1.0 |
0.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.0 |
13.84 |
10.3 |
2.33 |
4.420601 |
156 |
327 |
47 |
11.8 |
1.02 |
38.3 |
12.4 |
2.69 |
8.22 |
4.98 |
85.0 |
0.72 |
2.27 |
0.317181 |
3.51 |
1.34 |
5.7 |
12.0 |
43.0 |
150.0 |
102.0 |
9 |
0 |
0 |
0 |
2 |
1 |
| 4 |
26 |
1 |
2 |
66.0 |
0.0 |
2 |
3 |
3 |
0.0 |
0.0 |
0.0 |
1 |
1 |
0 |
0 |
0 |
1 |
0.0 |
4.73 |
11.6 |
2.56 |
4.531250 |
142 |
281 |
40 |
12.5 |
1.08 |
26.1 |
18.2 |
2.49 |
6.36 |
4.02 |
95.0 |
0.82 |
2.94 |
0.278912 |
4.14 |
1.67 |
5.9 |
22.0 |
17.0 |
147.0 |
75.0 |
3 |
0 |
0 |
0 |
2 |
1 |
| 5 |
27 |
0 |
2 |
74.0 |
1.0 |
3 |
3 |
3 |
0.0 |
0.0 |
0.0 |
1 |
0 |
0 |
1 |
0 |
0 |
0.0 |
6.18 |
5.07 |
0.54 |
9.388889 |
110 |
152 |
32 |
12.3 |
1.06 |
26.7 |
16.1 |
3.33 |
10.40 |
7.07 |
80.3 |
1.11 |
2.71 |
0.409594 |
4.20 |
0.97 |
6.7 |
19.0 |
22.0 |
125.0 |
80.0 |
5 |
0 |
0 |
0 |
0 |
0 |
| 6 |
30 |
1 |
2 |
70.0 |
1.0 |
1 |
1 |
1 |
1.0 |
1.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.0 |
8.5 |
6.19 |
1.61 |
3.844720 |
136 |
258 |
40 |
11.3 |
0.97 |
31.4 |
14.1 |
4.29 |
5.98 |
4.40 |
54.3 |
1.13 |
3.59 |
0.314763 |
5.28 |
1.00 |
5.4 |
15.0 |
15.0 |
147.0 |
89.0 |
2 |
0 |
0 |
0 |
0 |
0 |
| 7 |
34 |
0 |
1 |
58.0 |
0.0 |
1 |
1 |
1 |
1.0 |
1.0 |
0.0 |
1 |
1 |
0 |
0 |
0 |
0 |
0.0 |
9.48 |
6.85 |
1.73 |
3.959538 |
152 |
203 |
47 |
10.1 |
0.87 |
31.2 |
12.7 |
4.43 |
10.46 |
8.37 |
72.8 |
1.42 |
3.55 |
0.400000 |
4.96 |
1.51 |
8.6 |
13.4 |
43.0 |
160.0 |
105.0 |
4 |
0 |
0 |
0 |
1 |
0 |
| 8 |
36 |
1 |
2 |
65.0 |
0.0 |
1 |
1 |
1 |
0.0 |
0.0 |
1.0 |
0 |
0 |
0 |
0 |
0 |
1 |
0.0 |
15.6 |
11.52 |
3.06 |
3.764706 |
142 |
256 |
43 |
11.2 |
0.96 |
31.4 |
11.6 |
4.63 |
7.32 |
4.75 |
50.6 |
1.08 |
2.02 |
0.534653 |
3.52 |
1.02 |
6.5 |
21.0 |
15.0 |
110.0 |
76.0 |
11 |
0 |
0 |
1 |
3 |
1 |
| 9 |
38 |
0 |
2 |
82.0 |
1.0 |
3 |
3 |
3 |
1.0 |
0.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
10.31 |
8.12 |
1.24 |
6.548387 |
125 |
247 |
37 |
11.3 |
0.97 |
31.4 |
12.5 |
4.79 |
6.25 |
4.88 |
72.2 |
1.65 |
2.56 |
0.644531 |
4.21 |
0.77 |
5.6 |
18.7 |
25.0 |
220.0 |
104.0 |
4 |
0 |
0 |
0 |
1 |
0 |
分离 data 和 label
label_1 = df.columns[-3]
label_2 = df.columns[-1]
label1_data = df[label_1]
label2_data = df[label_2]
data = df[[column for column in df.columns if column not in [label_1, label_2]]]
data.drop(['index'], axis=1, inplace=True)
data.drop(['90dmRS'], axis=1, inplace=True)
data.head(10)
|
Sex |
Medication before thrombolytic therapy |
Age |
Age.1 |
Periventricular White Matter |
Deep White Matter |
The degree of WMH |
Smoking |
Drinking |
AtrialFibrillation |
Hypertension |
Diabetes |
Hyperlipidemia |
Coronary heart disease |
Heart failure |
Stroke |
TIA |
WBC |
N |
L |
NLR |
HB |
PLT |
PCV |
PT |
INR |
APTT |
TT |
Fibrinogen |
Emergency blood sugar |
Fasting blood glucose |
Creatinine |
HDL |
LDL |
HDL/LDL比值 |
Cholesterol |
Triglyceride |
HBLAC |
HCY |
DNT |
sBP |
dBP |
Baseline NIHSS score |
Hemorrhagic Transformation(HT) |
Early neurological deterioration (END) |
| 0 |
0 |
2 |
58.0 |
0.0 |
2 |
1 |
2 |
1.0 |
1.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
8.4 |
5.44 |
2.35 |
2.310000 |
141 |
266 |
38 |
10.7 |
0.92 |
34.5 |
14.9 |
3.52 |
5.68 |
5.11 |
66.2 |
0.92 |
3.15 |
0.290000 |
4.27 |
1.99 |
4.6 |
11.0 |
25.0 |
177.0 |
90.0 |
8 |
0 |
0 |
| 1 |
0 |
2 |
53.0 |
0.0 |
2 |
1 |
2 |
1.0 |
1.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
11.6 |
6.5 |
4.04 |
1.608911 |
145 |
259 |
45 |
9.9 |
0.86 |
30.6 |
15.3 |
3.34 |
6.38 |
6.13 |
74.0 |
1.46 |
4.54 |
0.321586 |
6.11 |
2.39 |
5.5 |
11.3 |
37.0 |
166.0 |
98.0 |
4 |
0 |
0 |
| 2 |
1 |
2 |
77.0 |
1.0 |
3 |
3 |
3 |
0.0 |
0.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
1 |
0.0 |
7.83 |
6.63 |
0.59 |
11.237288 |
103 |
225 |
30 |
9.2 |
0.80 |
28 |
14.9 |
4.82 |
1.76 |
7.11 |
107.9 |
1.19 |
1.03 |
1.155340 |
2.28 |
0.41 |
5.0 |
13.0 |
30.0 |
150.0 |
90.0 |
9 |
0 |
0 |
| 3 |
0 |
2 |
65.0 |
0.0 |
1 |
0 |
1 |
1.0 |
0.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.0 |
13.84 |
10.3 |
2.33 |
4.420601 |
156 |
327 |
47 |
11.8 |
1.02 |
38.3 |
12.4 |
2.69 |
8.22 |
4.98 |
85.0 |
0.72 |
2.27 |
0.317181 |
3.51 |
1.34 |
5.7 |
12.0 |
43.0 |
150.0 |
102.0 |
9 |
0 |
0 |
| 4 |
1 |
2 |
66.0 |
0.0 |
2 |
3 |
3 |
0.0 |
0.0 |
0.0 |
1 |
1 |
0 |
0 |
0 |
1 |
0.0 |
4.73 |
11.6 |
2.56 |
4.531250 |
142 |
281 |
40 |
12.5 |
1.08 |
26.1 |
18.2 |
2.49 |
6.36 |
4.02 |
95.0 |
0.82 |
2.94 |
0.278912 |
4.14 |
1.67 |
5.9 |
22.0 |
17.0 |
147.0 |
75.0 |
3 |
0 |
0 |
| 5 |
0 |
2 |
74.0 |
1.0 |
3 |
3 |
3 |
0.0 |
0.0 |
0.0 |
1 |
0 |
0 |
1 |
0 |
0 |
0.0 |
6.18 |
5.07 |
0.54 |
9.388889 |
110 |
152 |
32 |
12.3 |
1.06 |
26.7 |
16.1 |
3.33 |
10.40 |
7.07 |
80.3 |
1.11 |
2.71 |
0.409594 |
4.20 |
0.97 |
6.7 |
19.0 |
22.0 |
125.0 |
80.0 |
5 |
0 |
0 |
| 6 |
1 |
2 |
70.0 |
1.0 |
1 |
1 |
1 |
1.0 |
1.0 |
0.0 |
0 |
0 |
0 |
0 |
0 |
0 |
0.0 |
8.5 |
6.19 |
1.61 |
3.844720 |
136 |
258 |
40 |
11.3 |
0.97 |
31.4 |
14.1 |
4.29 |
5.98 |
4.40 |
54.3 |
1.13 |
3.59 |
0.314763 |
5.28 |
1.00 |
5.4 |
15.0 |
15.0 |
147.0 |
89.0 |
2 |
0 |
0 |
| 7 |
0 |
1 |
58.0 |
0.0 |
1 |
1 |
1 |
1.0 |
1.0 |
0.0 |
1 |
1 |
0 |
0 |
0 |
0 |
0.0 |
9.48 |
6.85 |
1.73 |
3.959538 |
152 |
203 |
47 |
10.1 |
0.87 |
31.2 |
12.7 |
4.43 |
10.46 |
8.37 |
72.8 |
1.42 |
3.55 |
0.400000 |
4.96 |
1.51 |
8.6 |
13.4 |
43.0 |
160.0 |
105.0 |
4 |
0 |
0 |
| 8 |
1 |
2 |
65.0 |
0.0 |
1 |
1 |
1 |
0.0 |
0.0 |
1.0 |
0 |
0 |
0 |
0 |
0 |
1 |
0.0 |
15.6 |
11.52 |
3.06 |
3.764706 |
142 |
256 |
43 |
11.2 |
0.96 |
31.4 |
11.6 |
4.63 |
7.32 |
4.75 |
50.6 |
1.08 |
2.02 |
0.534653 |
3.52 |
1.02 |
6.5 |
21.0 |
15.0 |
110.0 |
76.0 |
11 |
0 |
0 |
| 9 |
0 |
2 |
82.0 |
1.0 |
3 |
3 |
3 |
1.0 |
0.0 |
0.0 |
1 |
0 |
0 |
0 |
0 |
0 |
0.0 |
10.31 |
8.12 |
1.24 |
6.548387 |
125 |
247 |
37 |
11.3 |
0.97 |
31.4 |
12.5 |
4.79 |
6.25 |
4.88 |
72.2 |
1.65 |
2.56 |
0.644531 |
4.21 |
0.77 |
5.6 |
18.7 |
25.0 |
220.0 |
104.0 |
4 |
0 |
0 |
训练
def train(model, dataset, labelset):
x_train, x_test, y_train, y_test = train_test_split(dataset.values
, labelset.values
, test_size=0.2
, train_size=0.8
, shuffle=True
, stratify=labelset)
model.fit(x_train, y_train)
score = model.score(x_test, y_test)
accs = cross_val_score(model, dataset.values, labelset.values, verbose=0)
print(f'validation acc is: {score}')
print(f'cross validation accs are: {accs}')
y_pre = model.predict(x_test)
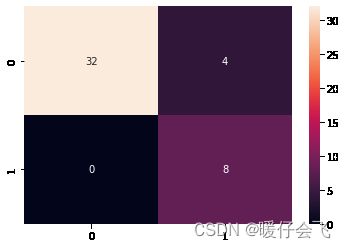
metri = confusion_matrix(y_test, y_pre)
sns.heatmap(metri, annot=True)
plt.show()
训练结果 & 混淆矩阵
svc1 = SVC(class_weight='balanced', kernel='linear')
train(svc1, data, label1_data)
validation acc is: 0.8863636363636364
cross validation accs are: [0.90909091 0.69767442 0.76744186 0.88372093 0.88372093]

svc2 = SVC(class_weight='balanced', kernel='linear')
train(svc2, data, label2_data)
validation acc is: 0.8863636363636364
cross validation accs are: [0.81818182 0.6744186 0.72093023 0.76744186 0.76744186]

lr1 = LogisticRegression(class_weight='balanced', max_iter=10000)
train(lr1, data, label1_data)
validation acc is: 0.9090909090909091
cross validation accs are: [0.81818182 0.6744186 0.76744186 0.88372093 0.90697674]
lr2 = LogisticRegression(class_weight='balanced', max_iter=10000)
train(lr2, data, label2_data)
validation acc is: 0.8863636363636364
cross validation accs are: [0.79545455 0.69767442 0.74418605 0.6744186 0.81395349]

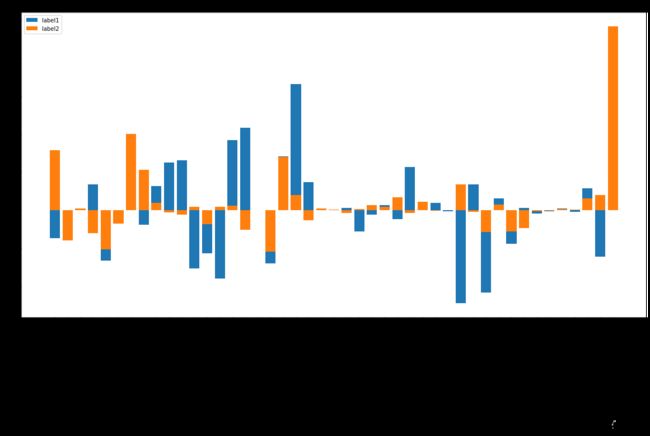
各种 feature 的重要性
def make_coef_dictNdf(data_columns, coef):
name_influence_dic = {string: imp for string, imp in zip(data_columns, coef.squeeze())}
name_influence_df = pd.DataFrame(data=name_influence_dic, index=['influence']).T
return name_influence_dic, name_influence_df
def write(filename, name_df_dic):
writer = pd.ExcelWriter(filename)
for k,v in name_df_dic.items():
v.to_excel(writer, sheet_name=k)
writer.save()
writer.close()
def plot(figsize, name_influence_df_lst, img_label_lst, title):
plt.figure(figsize=figsize)
for i in range(len(name_influence_df_lst)):
df = name_influence_df_lst[i]
plt.bar(x=df.index, height=df['influence'],label=img_label_lst[i])
plt.title(title)
plt.legend()
plt.xticks(rotation=90)
lr1_dic, lr1_df = make_coef_dictNdf(data.columns, lr1.coef_)
lr2_dic, lr2_df = make_coef_dictNdf(data.columns, lr2.coef_)
svc1_dic, svc1_df = make_coef_dictNdf(data.columns, svc1.coef_)
svc2_dic, svc2_df = make_coef_dictNdf(data.columns, svc2.coef_)
write("逻辑回归.xlsx", {'label1': lr1_df, 'label2': lr2_df})
write("SVM.xlsx", {'label1': svc1_df, 'label2': svc2_df})
plot((20,10), [lr1_df, lr2_df], ['label1', 'label2'], 'lr')
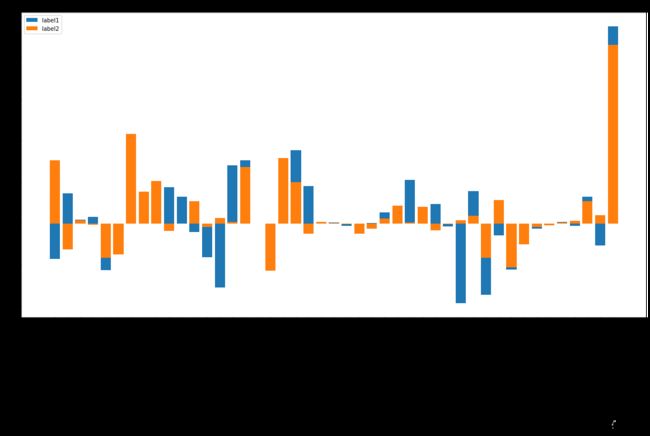
plot((20,10), [svc1_df, svc2_df], ['label1', 'label2'], 'svm')