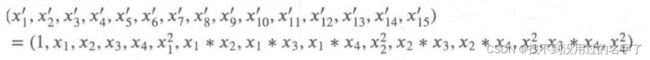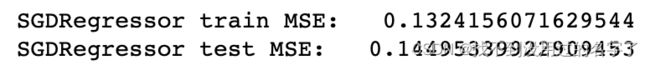【啃书系列】机器学习(1)
【啃书系列】——机器学习
第一部分 工业蒸汽量预测
1、赛题模型总括【机器学习】
(1)回归预测模型
输入:实值或者是离散数
输出:一个连续值域上的任意值
分类:
- 多元回归问题:多个输入变量的回归问题
- 时间序列预测问题:输入变量按时间排序的回归问题
(2)分类预测模型
输入:实值或者是离散数
输出:将实例分为两个或多个类中的一个
分类:
- 二类分类问题/二元分类问题
- 多类别分类问题
2、过程
- 划分数据集
- 缺失值补充
- 异常值处理
- 处理特征(特征工程)
- 特征降维
- 模型训练
- 模型验证
- 特征优化
- 模型融合
3、解题思路
需要根据V0~V37一共38个特征变量来预测蒸汽量的数值,其预测值为连续兴数值变量,所以是回归预测求解。模型可以选用:
- 线性回归【Linear Regression】
- 岭回归【Ridge Regression】
- LASSO回归
- 决策树回归【Decision Tree Regressor】
- 梯度提升树回归【Gradient Boosting Decision Tree Regressor】
4、数据分析
(1)数据集【百度网盘满了,懒得删了,加上不想买会员,如果想要可以私聊我】
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
train_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file,sep='\t',encoding='utf-8')
test_data = pd.read_csv(test_data_file,sep='\t',encoding='utf-8')
(2)变量识别
#1、看值
train_data.head()
test_data.head()
#2、看形状
train_data.shape
test_data.shape
#3、看训练集和测试集的占比
train_data.shape[0]/(train_data.shape[0]+test_data.shape[0])
test_data.shape[0]/(train_data.shape[0]+test_data.shape[0])
#4、查看数据基本信息:看有无缺失值(说明没有缺失值)和数据类型
train_data.info()
test_data.info()
(3)划分数据集
#划分数据集:X和label,train和test[大赛给的test是没有标签的,我们需要在train数据集里面划分我们自己的train和test]
from sklearn.model_selection import train_test_split
X = train_data.drop('target',axis=1)
y = train_data.target
X.shape,y.shape
X_train,X_test,y_train,y_test = train_test_split(X,y,test_size=0.2,shuffle=True)#80%做训练集,20%做测试集
X_train.shape,X_test.shape,y_train.shape,y_test.shape
(4)变量分析
- 单变量分析
- 连续型变量:统计数据的中心分布趋势和变量的分布
- 类别型变量:使用频次或占比表示每个类别的分布情况,对应的衡量指标分别是类别变量的频次(次数)和频率(占比),可以用柱形图来表示可视化分布情况。
- 双变量分析
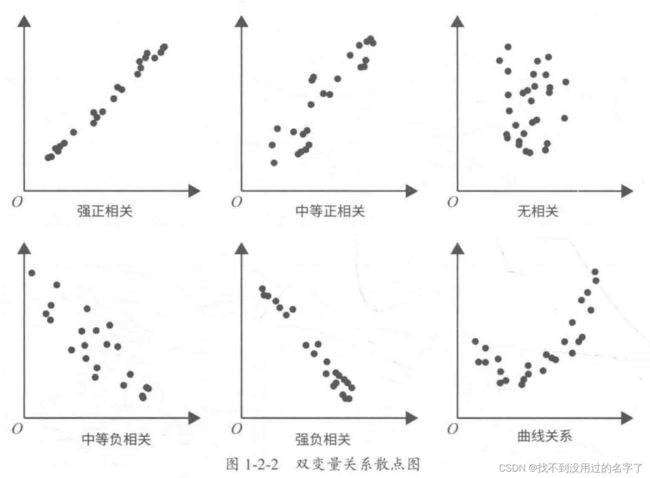
- 连续型 VS 连续型
(1) 【直观】绘制散点图
(2) 【量化】计算相关性
C o r r e l a t i o n = C o n v a r i a n c e ( X , Y ) V a r ( X ) ∗ V a r ( Y ) Correlation = \frac{Convariance(X,Y)}{\sqrt{Var(X)*Var(Y)}} Correlation=Var(X)∗Var(Y)Convariance(X,Y)
相关性系数的取值区间为 [1, 1] 。
一般来说,在取绝对值后 ,0-0.09 为没有相关性, 0.1-0.3 为弱相关, 0.3-0.5 为中相关, 0.5-1.0 为强相关。
代码如下
import nurnpy as np
X = np.array((65 , 72, 78 , 65 , 72 , 70 , 65 , 68])
Y=np . array([72 , 69 , 79 , 69 , 84 , 75 , 60 , 73])
np.corrcoef(X, Y)
- 类别型 VS 类别型
(1) 双向表
(2) 堆叠柱状图
(3) 卡方检验:主要用于两个和两个以上样本率(构成比)及两个二值型离散变量的关联性分析,即比较伦理频次与实际频次的吻合程度或拟合程度
from sklearn.da asets import load_ iris
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
iris= load_iris()
X, y = iris.data, iris.target
chiValues = chi2(X, y)
X new = Select.KBest(chi2, k=2).fit_transform(X, y)
pd.set_option ('display.max_columns', 10)
pd.set_option ('display.max_rows', 10)
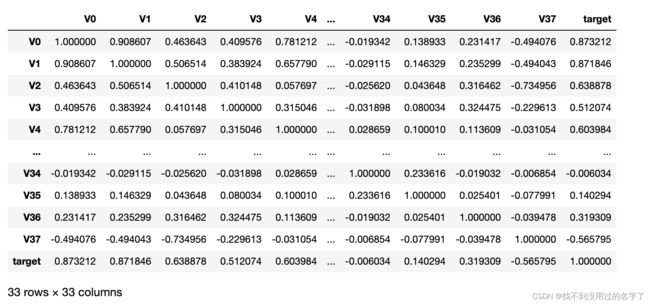
data_trainl =train_data.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
train_corr = data_trainl.corr()
train_corr
ax = plt.subplots(figsize= (20, 16)) #调整画布大小
ax= sns.heatmap(train_corr,vmax=.8 , square=True, annot=True)#画热力图
(5)缺失值处理【没有缺失值,没进行】
-
- 一般填充是用该变量下所有非缺失值的平均值或中值来补全缺失值。
-
- 相似样本填充是利用具有相似特征的样本的值或者近似值进行填充。
- 预测模型填充
在这种情况下,会把数据集分为两份
一份是没有缺失值的 ,用作训练集;
另一份是有缺失值的 ,用作测试集 。
这样缺失的变量就是预测目标,此时可以使用回归、分类等方法来完成填充。
(6)异常值处理
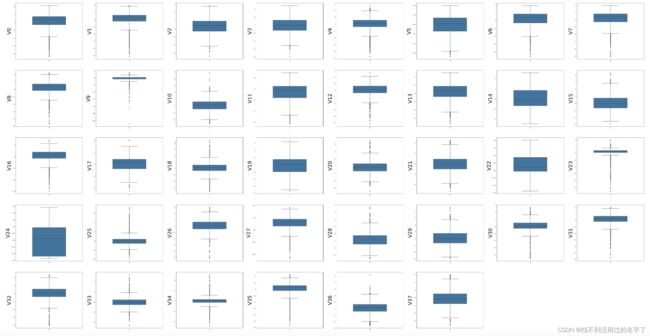
操作1. 异常值检测
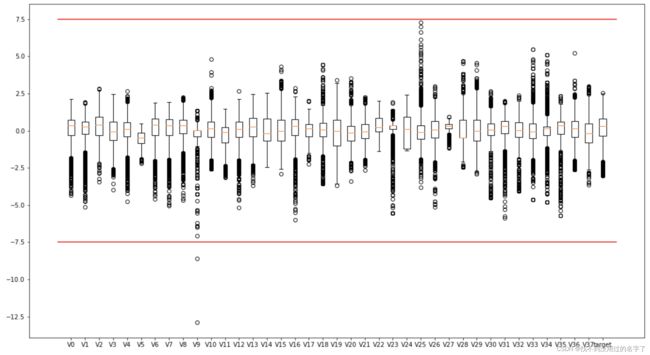
#所有变量的箱型图
column = train_data.columns.tolist()[:39] #列表头
fig = plt.figure(figsize=(80,60),dpi=75) #指定绘图对象的宽度和高度
for i in range(38):
plt.subplot(7,8,i+1) #8列子图
sns.boxplot(data=train_data[column[i]],width=0.5)
plt.ylabel(column[i],fontsize=36)#显示的纵坐标的大小
plt.show()
#另一种画法
plt.figure(figsize=(18,10))
plt.boxplot(x=train_data.values,labels=train_data.columns)
plt.hlines([-7.5,7.5],0,40,colors='r')#0,40是这条线的长度
plt.show()
-
- 直方图
-
- 散点图
操作2.寻找具体异常值
#异常值的处理
def find_outliers(model,X,y,sigma=3):
try:#用模型来预测y值
y_pred = pd.Series(model.predict(X),index=y.index)
except:#如果预测失败,尝试先调整模型
model.fit(X,y)
y_pred = pd.Series(model.predict(X),index=y.index)
resid = y-y_pred #模型的预测值和真实值相减
mean_resid = resid.mean()#均值
std_resid = resid.std()#方差
z = (resid-mean_resid)/std_resid
outliers = z[abs(z)>sigma].index#异常值
print('R2=',model.score(X,y))#用来度量样本回归线的拟合优度=回归平方和/总平方和
print('mse=',mean_squared_error(y,y_pred))
print('-----------------------------------------')
print('mean of residuals:',mean_resid)
print('std of residuals:',std_resid)
print('-----------------------------------------')
print((len(outliers),'outliers:'))
print(outliers.tolist())
plt.figure(figsize=(15,5))
ax_131 = plt.subplot(1,3,1)
plt.plot(y,y_pred,'.')#中间的蓝点
plt.plot(y.loc[outliers],y_pred.loc[outliers],'ro')#outlier 的红点
plt.legend(['Accepted','Outlier'])#顺序要和上面的对应
plt.xlabel('y')
plt.ylabel('y_pred')
ax_132 = plt.subplot(1,3,2)
plt.plot(y,y-y_pred,'.')
plt.plot(y.loc[outliers],y.loc[outliers]-y_pred.loc[outliers],'ro')
plt.legend(['Accepted','Outlier'])
plt.xlabel('y')
plt.ylabel('y-y_pred')
ax_133 = plt.subplot(1,3,3)
z.plot.hist(bins=50,ax=ax_133)#hist直方图
z.loc[outliers].plot.hist(color='r',bins=50,ax=ax_133)
plt.legend(['Accepted','Outlier'])
plt.xlabel('z')
plt.savefig('outliers.png')
return outliers
#用数据预测y y值与均值之差 超过多少个方差的 视为异常值
outliers = find_outliers(Ridge(),X_train,y_train,sigma=3)
操作2. 异常值处理
-
- 删除
-
- 转换:数据转换可以消除异常值 如对数据取对数会减轻由极值引起的变化
-
- 填充:像处理缺失值一样,我们可以对异常值进行修改,如使用平均值、中值或其他的一些填补方法。在填补之前 需要分析异常值是自然形成的,还是人为造成的。如果是人为造成的,则可以进行填充处理,如使用预测模型填充
-
- 区别对待 :如果存在大量的异常值,则应该在统计模型中区别对待。其中一个方法是将数据分为两个不同的组:异常值归为一组,非异常值归为一组,且两组分别建立模型,然后最终将两组的输出合并。
这里采取的是删除法
- 区别对待 :如果存在大量的异常值,则应该在统计模型中区别对待。其中一个方法是将数据分为两个不同的组:异常值归为一组,非异常值归为一组,且两组分别建立模型,然后最终将两组的输出合并。
#1、删除异常值(横着删)
X_train = X_train.drop(outliers)
y_train = y_train.drop(outliers)
X_train.shape,y_train.shape
#2、删除异常值(竖着删)
#针对每一个特征向量删掉样本异常点
for f in X_train.columns:
X_ = X_train.drop(f,axis=1)#把特征值“V0”删掉
y_ = X_train[f]#特征值“V0”
outliers = find_outliers(Ridge(),X_,y_,sigma=4)
X_train = X_train.drop(outliers)
y_train = y_train.drop(outliers)
X_train.shape,y_train.shape
5、特征工程
1、数据预处理
- 数据采集
- 数据清洗
- 数据采样:
数据在采集、清洗过以后 正负样本是不均衡的,故要进行数据采样。采样的方法有随机采样和分层抽样。但由千随机采样存在隐患,可能某次随机采样得到的数据很不均匀,因此更多的是根据特征进行分层抽样》
正负样本不平衡的处理方法 -
- 正样本>负样本,且量都特别大的情况;采用下采样(downsampling)的方法。
-
- 正样本>负样本,且量不大的情况,可采用以下方法采集更多的数据:上采样(oversampling),比如图像识别中的镜像和旋转;修改损失函数(loss function)设置样本权重。
2、特征处理
- 标准化
依照特征矩阵的列处理数据 ,即通过求标准分数的方法,将特征转换为标准正态分布,并和整体样本分布相关。每个样本点都能对标准化产生影响。
标准化需要计算特征的均值和标准差,公式如下
x ′ = x − X ‾ S x^{'}=\frac{x-\overline{X}}{S} x′=Sx−X
from sklearn.preprocessing import StandardScaler
StandardScaler().fit_transform(iris.data)
- 区间缩放法
区间缩放法的思路有多种,常见的一种是利用两个最值(最大值和最小值)进行缩放。
x ′ = x − M i n M a x − M i n x^{'}=\frac{x-Min}{Max-Min} x′=Max−Minx−Min
from sklearn.preprocessing import MinMaxScaler
MinMaxScaler().fit_transform(iris.data)
- 归一化
归一化是将样本的特征值转换到同 量纲下,把数据映射到[0,1] 或者[a,b] 区间内,由于其仅由变量的极值决定,因此区间缩放法是归一化的一种。
归一区间会改变数据的原始距离、分布和信息,但标准化般不会。
规则为L2的归一化公式如下:
x ′ = x ∑ j m x [ j ] 2 x^{'}=\frac{x}{\sqrt{\sum_j^m{x[j]}^2}} x′=∑jmx[j]2x
from sklearn.preprocessing import Normalizer
Normalizer().fit_transform(iris.data)
归一化与标准化的使用场景:
- 如果对输出结果范围有要求,则用归一化。
- 如果数据较为稳定,不存在极端的最大值或最小值,则用归一化。
- 如果数据存在异常值和较多噪声,则用标准化,这样可以通过中心化间接避免异常值和极端值的影响。
- 支持向量机 (Support Vector Machine, SVM)、K近邻 (K-Nearest Neighbor, KNN)、主成分分析 (Principal Component Analysis, PCA) 等模型都必须进行归一化或标准化操作。
- 定量特征二值化
定量特征二值化的核心在于设定一个阙值,大于阙值的赋值为1,小于等于阙值的赋值为0.
from sklearn.preprocessing import Binarizer
Binarizer(threshold=3).fit_transform(iris.data)
- 定性特征哑编码
哑变量,也被称为虚拟变量,通常是人为虚设的变量,取值为0或1,用来反映某个变量的不同属性。将类别变量转换为哑变量的过程就是哑编码。而对于有n个类别属性的变量,通常会以1个类别特征为参照,产生n-1个哑变量。
引入哑变量的目的是把原本不能定量处理的变量进行量化,从而评估定性因素对因变量的影响。
from sklearn.preprocessing import OneHotEncoder
OneHotEncoder(categories='auto').fit_transform(iris.target.reshape((-1,1)))
- 缺失值处理
当数据中存在缺失值时,用 Pandas 读取后特征均为 NaN, 表示数据缺失,即数据未知.
from numpy import vstack,array,nan
from sklearn.impute import SimpleImputer
#缺失值处理,返回值为处理缺失值后的数据
#参数 missing_value 为缺失俏的表示形式,默认为 NaN
#参数 strategy 为缺失值的填充方式,默认为 mean (均值)
SimpleImputer().fit_transform(vstack((array([nan,nan,nan,nan]),iris.data)))
- 数据转换
from sklearn.preprocessing import PolynomialFeatures
PolynomialFeatures().fit_transfor(iris.data)
-
- 对数变换
基于单变元函数的数据转换可以使用一个统一的方法完成。
- 对数变换
from numpy import loglp
from sklearn.preprocessing import FunctionTransformer
FunctionTransformer(loglp,validate=False).fit_transform(iris.data)
针对本题代码:
#(1)用sklearn归一化【建议】
from sklearn import preprocessing
features_columns = list(X_train.columns)
min_max_scaler = preprocessing.MinMaxScaler()#缩放
min_max_scaler = min_max_scaler.fit(X_train[features_columns])
X_train_scaler = pd.DataFrame(min_max_scaler.transform(X_train[features_columns]))
X_test_scaler = pd.DataFrame(min_max_scaler.transform(X_test[features_columns]))#自己分的测试集
X_test_data_scaler = pd.DataFrame(min_max_scaler.transform(test_data[features_columns]))#大赛给的测试集
#(2)标准化变换过程
#法二:使用sklearn实现Yeo-Johnson
import numpy as np
from sklearn.preprocessing import PowerTransformer
pt = PowerTransformer()#这里method默认是Yeo-Johnson
pt.fit(X_train_scaler)
X_train_s_bc = pt.transform(X_train_scaler)
X_test_s_bc = pt.transform(X_test_scaler)
X_test_data_s_bc = pt.transform(X_test_data_scaler)
X_train_s_bc = pd.DataFrame(X_train_s_bc)
X_test_s_bc = pd.DataFrame(X_test_s_bc)
X_test_data_s_bc = pd.DataFrame(X_test_data_s_bc)
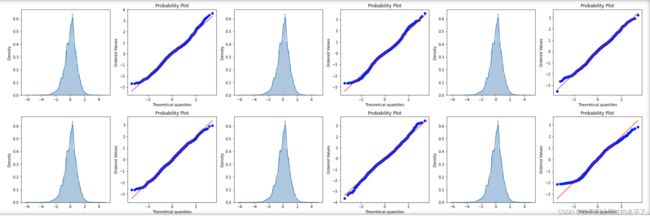
#(3)查看目前的数据状况(之前也要查看滴)
#qq图【散点图】是数据分位数(蓝色那条)与正态分布分位数(红色那条)
#直方图:数据的分布图
train_cols = 6
train_rows = len(X_train_s_bc.columns)
plt.figure(figsize=(4*train_cols,4*train_rows))
i=0
for col in X_train_s_bc.columns:
i += 1
ax = plt.subplot(train_rows,train_cols,i)
sns.distplot(X_train)#直方图
i += 1
ax = plt.subplot(train_rows,train_cols,i)
res = stats.probplot(X_train_s_bc[col],plot=plt)#qq图
plt.tight_layout()
plt.show()
3、特征降维
-
- VarianceThreshold【方差选择法 】
先计算各个特征的方差,然后根据阈值选择方差大于阈值的特征
- VarianceThreshold【方差选择法 】
from sklearn.feature_selection import VarianceThreshold
from sklearn.datasets import load_iris
iris = load_iris()
#方差选择法,返回值为特征选择后的数据
#参数threshold为方差的阈值
VarianceThreshold(threshold=3).fit_transform(iris.data)
-
- SelectKBest
将可选相关系数、卡方检验或最大信息系数作为得分计算的方法。
- SelectKBest
-
-
- 相关系数法。使用相关系数法,先要计算各个特征对目标值的相关系数及相关系数的P值,然后根据阈值筛选特征。
-
import numpy as np
from sklearn.datasets import load_iris
iris = load_iris()
from array import array
from sklearn.feature_selection import SelectKBest
from scipy.stats import pearsonr
#选择K个最好的特征 返回选择特征后的数据
#第一个参数为计算评估特征的函数,该函数输入特征矩阵和目标向量,输出二元组(评分,P值)的数组,数组第i项为第i个特征的评分和P值。在此定义为计算相关系数
#参数k为选择的特征个数
SelectKBest(lambda X,Y:np.array(list(map(lambda x:pearsonr(x, Y),X.T))).T[0],k=2).fit_transform(iris.data,iris.target)
-
-
- 卡方检验。经典的卡方检验是检验定性自变量与定性因变量的相关性。假设自变量有N种取值,因变量有 M 种取值,考虑自变量等于 i 且因变量等于 j 的样本频数的观察值与期望的差距,构建统计量:
x 2 = ∑ ( A − E ) 2 E x^2=\sum\frac{(A-E)^2}{E} x2=∑E(A−E)2
- 卡方检验。经典的卡方检验是检验定性自变量与定性因变量的相关性。假设自变量有N种取值,因变量有 M 种取值,考虑自变量等于 i 且因变量等于 j 的样本频数的观察值与期望的差距,构建统计量:
-
from sklearn datasets import load_iris
iris = load_iris()
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
#选择k个最好的特征,返回选择特征后的数据
SelectKBest(chi2, k=2) fit_transform(iris.data,iris.target)
-
-
- 最大信息系数法。经典的互信息也是评价定性自变量与定性因变量相关性的方法。
互信息计算公式如下:
I ( X ; Y ) = ∑ x ∈ y ∑ y ∈ x p ( x , y ) l o g p ( x , y ) p ( x ) p ( y ) I(X;Y)=\sum_{x \in y}\sum_{y \in x}p(x,y)log\frac{p(x,y)}{p(x)p(y)} I(X;Y)=∑x∈y∑y∈xp(x,y)logp(x)p(y)p(x,y)
- 最大信息系数法。经典的互信息也是评价定性自变量与定性因变量相关性的方法。
-
import numpy as np
from sklearn.feature.selection import SelectKBest
from minepy import MINE
#由于 MINE 的设计不是函数式的,因此需要定义 mic 方法将其转换为函数式,返回一个二元组,二元组的第2项设置成固定的P值,为0.5
def mic(x, y):
m =MINE()
m.compute_score(x y)
return (m.mic(), 0.5)
#选择k个最好的特征,返回特征选择后的数据
SelectKBest(lambda X, Y: np.array(list(map(lambda x: mic(x, Y), X.T))).T[0],k=2).fit_transform(iris.data, iris. target)
-
- RFE【递归消除特征法】
使用一个基模型来进行多轮训练,每轮训练后,消除若干权值系数的特征,再基于新的特征集进行下一轮训练。
- RFE【递归消除特征法】
from sklearn.feature_selection import RFE
from sklearn.linear_model import LogisticRegression
#递归特征消除法,返回特征选择后的数据
#参数 estimator 为基模型
#参数 n_features_to_select 为选择的特征个数
RFE(estimator=LogisticRegression(multi_class='auto',solver= 'lbfgs',max_iter=5OO),n_features_to_select=2).fit_transform(iris.data,iris.target)
-
- SelectFromModel【基于模型的特征选择法】、
主要采用基于模型的特征选择法,常见的有基于惩罚项的特征选择法和基于树模型的特征选择法。
- SelectFromModel【基于模型的特征选择法】、
-
-
- 基千惩罚项的特征选择法使用带惩罚项的基模型,除了能筛选出特征,也进行了降维
-
from sklearn.feature_selection import SelectFromModel
from sklearn.linear_model import LogisticRegression
#将带 Ll 惩罚项的逻辑同归作为基模型的特征选择
SelectFromModel(
LogisticRegression(penalty='l2',C=0.1,solver='lbfgs',multi_class='auto')).fit_transform(iris.data,iris.target)
-
-
- 基于树模型的特征选择法。在树模型中, GBDT ( Gradient Boosting Decision Tree, 梯度提升迭代决策树)也可用来作为基模型进行特征选择。
-
from sklearn.feature_selection import SelectFromModel
from sklearn.ensemble import GradientBoostingClassifier
#将 GBDT 作为基模型的特征选择
SelectFromModel(GradientBoostingClassifier()).fit_transform(iris.data,iris.target)
这道题的代码
#统一特征
X_train_s_bc.columns = test_data.columns
X_test_s_bc.columns = test_data.columns
X_test_data_s_bc.columns = test_data.columns
dist_cols = 6
dist_rows = len(test_data.columns)
plt.figure(figsize=(4*dist_cols,4*dist_rows))
i = 1
for col in test_data.columns:
ax = plt.subplot(dist_rows,dist_cols,i)
ax = sns.kdeplot(train_data[col],color='Red',shade=True)
ax = sns.kdeplot(test_data[col],color='Blue',shade=True)
ax.set_xlabel(col)
ax.set_ylabel('Frequency')
ax = ax.legend(['train','test'])
i += 1
plt.show()
![]()
-
-
- 基本一致
-
-
-
- 存在训练集与数据集分布不一致的特征 5 9 11 17 22 28
-
-
-
- 这些特征会导致模型的泛化能力变差 考虑删除该特征
-
X_train_s_bc_t = pd.concat([X_train_s_bc,y_train],axis = 1)#带个target
X_train_s_bc_kde = X_train_s_bc.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
X_test_s_bc_kde = X_test_s_bc.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
X_test_data_s_bc_kde = X_test_data_s_bc.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
data_train1 = X_train_s_bc_t.drop(['V5','V9','V11','V17','V22','V28'],axis=1)
-
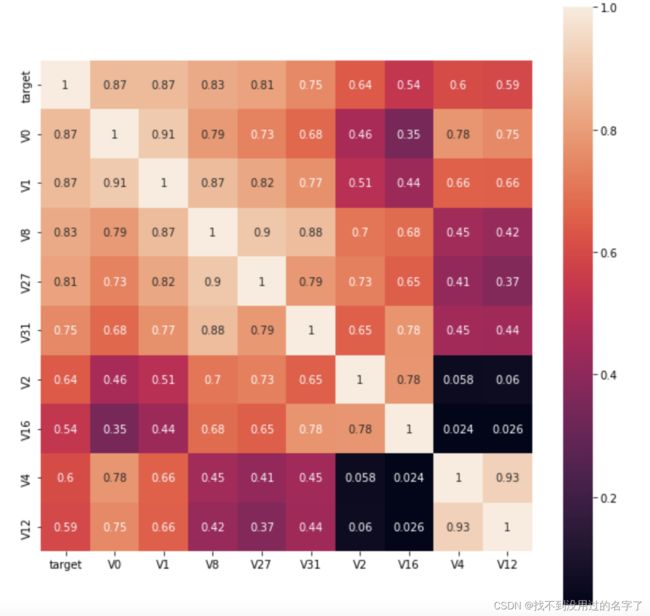
- 然后看哪些特征与target最相关
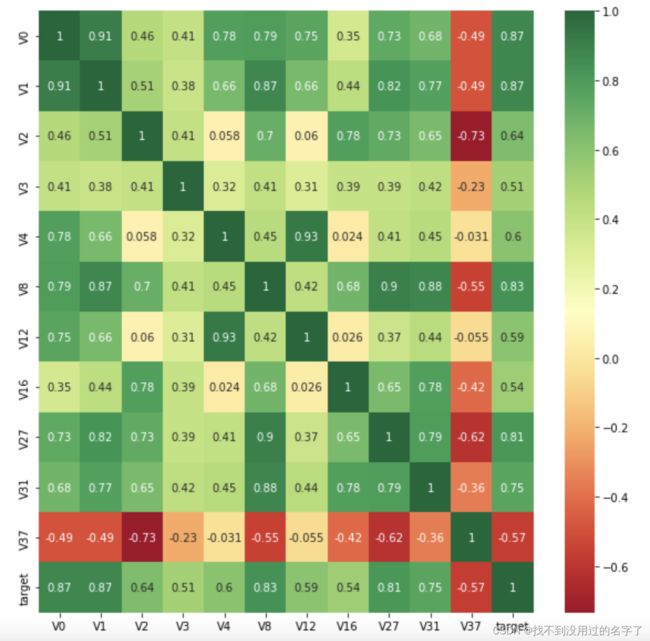
(1)画热力图
- 然后看哪些特征与target最相关
#k个与target最相关特征
k = 10
cols = train_corr.nlargest(k,'target')['target'].index
cm = np.corrcoef(train_data[cols].values.T)
hm = plt.subplots(figsize=(10,10)) #调整画布大小
hm = sns.heatmap(train_data[cols].corr(),annot=True,square=True)
plt.show()
#与target相关系数大于0.5的特征变量
threshold = 0.5
corrmat = train_data.corr()
top_corr_features = corrmat.index[abs(corrmat['target'])>threshold]
plt.figure(figsize=(10,10))
g = sns.heatmap(train_data[top_corr_features].corr(),annot=True,cmap='RdYlGn')
(2)相关性只是两个变量之间的关系,如果一个变量可以被其他三个变量解释呢,无法光靠相关性检验出来。用VIF。
from statsmodels.stats.outliers_influence import variance_inflation_factor
#多重共线性
new_numerical = X_train_s_bc_kde.columns.tolist()
X = X_train_s_bc_kde.values
VIF_list = [variance_inflation_factor(X,i) for i in range(X.shape[1])]
VIF_list
#一般0.9比较高
- 线性降维
-
- 主成分分析法(PCA)
主成分分析法是最常用的线性降维方法,主要原理是通过某种线性投影,将高维的数据映射到低维的空间中表示,并期望在所投影的维度上数据的方差最大,以此达到使用较少的数据维度来保留较多的原数据点特性的效果。
- 主成分分析法(PCA)
from sklearn decomposition import PCA
#主成分分析法,返回降维后的数据
#参数 n_components 为主成分的数目
PCA(n_components=2).fit_transform(iris.data)
-
- 线性判别分析法(LDA)
也叫做Fisher线性判别(FLD),是一种有监督的线性降维算法。与PCA尽可能多地保留数据信息不同,LDA的目标是使降低维度后的数据点尽可能地容易被区分,其利用了标签的信息。
假设原始线性数据为X,我们希望找到映射向量 a a a,使得 a X aX aX后的数据点能够保持以下两种性质:
- 线性判别分析法(LDA)
-
-
- 同类的数据点尽可能靠近
-
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis as LDA
#线性判别分析法,返回降维后的数据
#参数n_components为降维后的维数
LDA(n_components=2).fit_transform(iris.data, iris.target)
这道题的代码
#处理PCA降维
from sklearn.decomposition import PCA #主成分分析法
pca = PCA(n_components=0.95)
X_train_s_bc_kde_pca_95 = pca.fit_transform(X_train_s_bc_kde)
X_test_s_bc_kde_pca_95 = pca.transform(X_test_s_bc_kde)
X_test_data_s_bc_kde_pca_95 = pca.transform(X_test_data_s_bc_kde)
6、模型训练
1、回归及相关模型
- 概念:
用于在目标数量连续时预测所需目标数量的值。 - 线性回归模型:【感知机】
(1)假定因变量与自变量X呈线性相关,则可以采用线性模型找出自变量X和因变量Y的关系,以便预测新的自变量X的值 ,这就是线性回归模型。
from sklearn.model_selection import train_test_split
from sklearn.decomposition import PCA
pca = PCA (n_components=16)
features_columns = [col for col in train_data.columns if col not in ['target']]
train_data_scaler = pd.DataFrame(min_max_scaler.transform(train_data[features_columns]))
test_data_scaler = pd.DataFrame(min_max_scaler.transform(test_data[features_columns]))
new_train_pca_16 = pca.fit_transform(train_data_scaler.iloc[:, 0:-1])
new_test_pca_16 = pca.transform(test_data_scaler.iloc[:, 0:-1])
new_train_pca_16 = pd.DataFrame(new_train_pca_16)
new_test_pca_16 = pd.DataFrame(new_test_pca_16)
new_train_pca_16['target'] = train_data['target']
线性回归调用
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
new_train_pca_16 = new_train_pca_16.fillna(0) #采用 PCA 保留 16 维特征的数据
train= new_train_pca_16[new_test_pca_16.columns]
target = new_train_pca_16['target']
train_data, test_data,train_target,test_target = train_test_split(train,target,test_size=0.2,random_state=0)
clf = LinearRegression()
clf.fit(train_data,train_target)
test_pred = clf.predict(test_data)
score = mean_squared_error(test_target,clf.predict(test_data))
print("LinearRegression: ",score)
- K近邻回归模型
K近邻算法不仅可以用于分类,还可以用于回归 。通过找出某个样本的k个最近邻居,将这些邻居的某个(些)属性的平均值赋给该样本,就可以得到该样本对应属性的值。
from sklearn.neighbors import KNeighborsRegressor
clf = KNeighborsRegressor(n_neighbors=3)
clf.fit(train_data,train_target)
test_pred = clf.predict(test_data)
score = mean_squared_error(test_target,clf.predict(test_data))
print("KNeighborsRegressor: ",score)
#从 sklearn 算法库中导入决策树回归算法
from sklearn.tree import DecisionTreeRegressor
clf = DecisionTreeRegressor()
clf.fit(train_data,train_target)
test_pred = clf.predict(test_data)
score= mean_squared_error(test_target,clf.predict(test_data))
print("DecisionTreeRegressor: ", score)
- 集成学习回归模型
#从 sklearn 算法库中导入随机森林回归树模型
from sklearn.ensemble import RandomForestRegressor
clf= RandomForestRegressor(n_estimators=200) # 200 棵树模型
clf.fit(train_data,train_target)
test_pred = clf.predict(test_data)
score= mean_squared_error(test_target,clf.predict(test_data))
print ("RandomForestRegressor: ",score)
- LightGBM回归模型
# LGB 回们模型
clf= lgb.LGBMRegressor(learning_rate=0.01,max_depth=-1,n_estimators=5000,boosting_type='gbdt ',random_state=2019,objective='regression',)
#训练模型
clf.fit(X=train_data, y=train_target, eval_metric='MSE', verbose=50)
score = mean_squared_error(test_target,clf.predict(test_data))
print ("lightGbm: ", score)
7、模型验证
数据
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
from sklearn.linear_model import LinearRegression #线性回归
from sklearn.neighbors import KNeighborsRegressor # 近邻回归
from sklearn.tree import DecisionTreeRegressor #决策树回归
from sklearn.ensemble import RandomForestRegressor #随机森林回归
from sklearn.svm import SVR #支持向量回归
import lightgbm as lgb # LightGBM 模型
from sklearn.model_selection import train_test_split #切分数据
from sklearn.metrics import mean_squared_error #评价指标
from sklearn.linear_model import SGDRegressor
#读取数据
train_data_file = "./zhengqi_train.txt"
test_data_file = "./zhengqi_test.txt"
train_data = pd.read_csv(train_data_file, sep= '\t', encoding= 'utf-8')
test_data = pd.read_csv(test_data_file, sep='\t', encoding='utf-8')
#归一化处理
from sklearn import preprocessing
features_columns = [col for col in train_data.columns if col not in ['target']]
min_max_scaler = preprocessing.MinMaxScaler()
min_max_scaler = min_max_scaler.fit(train_data[features_columns])
train_data_scaler = min_max_scaler.transform(train_data[features_columns])
test_data_scaler = min_max_scaler.transform(test_data[features_columns])
train_data_scaler = pd.DataFrame(train_data_scaler)
train_data_scaler.columns= features_columns
test_data_scaler = pd.DataFrame(test_data_scaler)
test_data_scaler.columns = features_columns
train_data_scaler['target'] = train_data['target']
# PCA 方法降维
from sklearn.decomposition import PCA #主成分分析法
#保留 16 个上成分
pca= PCA(n_components=16)
new_train_pca_16 = pca.fit_transform(train_data_scaler.iloc[:,0:-1])
new_test_pca_16 = pca.transform(test_data_scaler)
new_train_pca_16 = pd.DataFrame(new_train_pca_16)
new_test_pca_16 = pd.DataFrame(new_test_pca_16)
new_train_pca_16['target'] =train_data_scaler['target']
#将数据切分为训练数据和验证数据
#保留16维特征并切分数据
new_train_pca_l6 = new_train_pca_16.fillna(0)
train= new_train_pca_l6[new_test_pca_16.columns]
target= new_train_pca_16['target']
#切分数据,训练数据为80% ,验证数据为20%
train_data, test_data, train_target, test_target = train_test_split(train,target, test_size=0.2, random_state=0)
1、欠拟合和过拟合
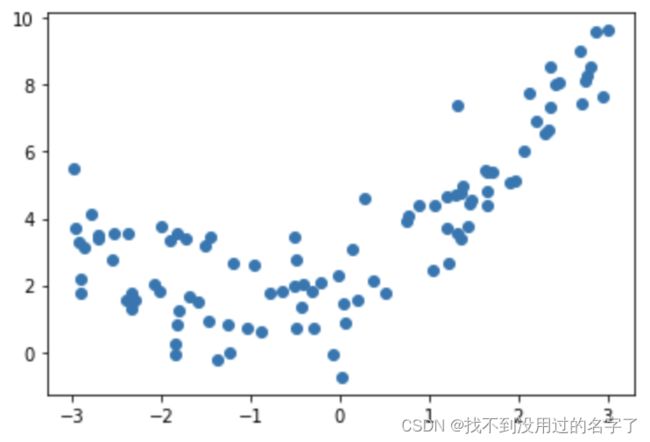
对于数据
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(666)
x = np.random.uniform(-3.0,3.0,size=100)
X = x.reshape(-1,1)
y = 0.5*x**2+x+2+np.random.normal(0,1,size=100)
plt.scatter(x,y)
plt.show()
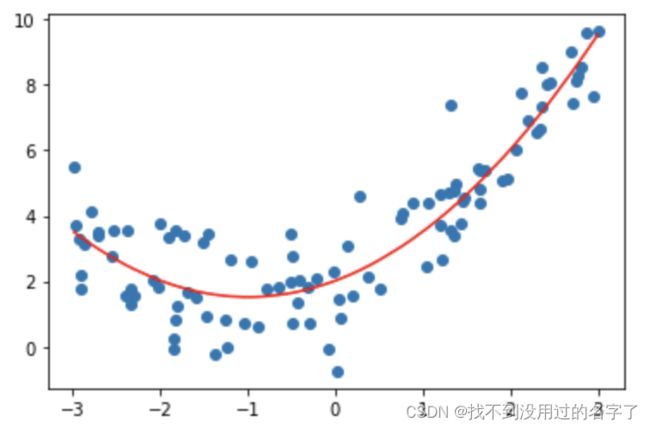
- 一元线性回归
(1)代码
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
lin_reg.fit(X,y)#训练
lin_reg.score(X,y)#评估模型性能
from sklearn.metrics import mean_squared_error
y_predict = lin_reg.predict(X)#预测
display(mean_squared_error(y,y_predict))
plt.scatter(x, y)
plt.plot(np.sort(x),y_predict[np.argsort(x)] , color='r')
plt.show()
#多项式拟合
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PolynomialFeatures
from sklearn.preprocessing import StandardScaler
def PolynomialRegression(degree):
return Pipeline ([('poly', PolynomialFeatures(degree=degree)),('std_scaler', StandardScaler()), ('lin reg ' , LinearRegression ())])
- 多项式参数设置为degree=2
poly2_reg = PolynomialRegression(degree=2)
poly2_reg.fit(X,y)
y2_predict = poly2_reg.predict(X)
#比较真实值和预测值的均方误差
display(mean_squared_error(y,y2_predict))
#拟合结果可视化
plt.scatter(x,y)
plt.plot(np.sort(x),y2_predict[np.argsort(x)],color='r')
plt.show()
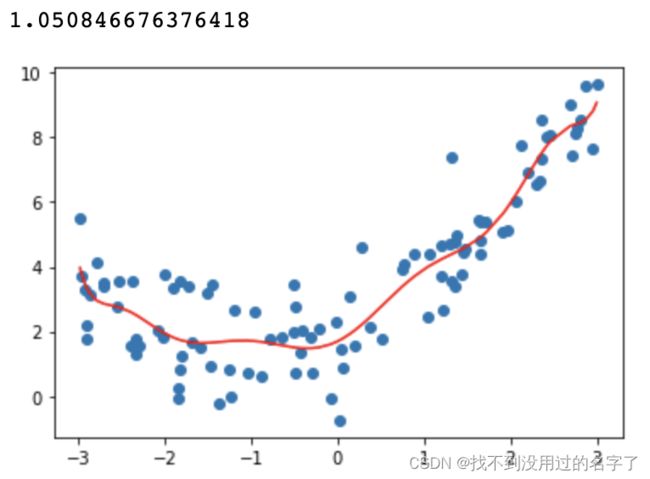
- 多项式参数设置为degree=10
poly10_reg = PolynomialRegression(degree=10)
poly10_reg.fit(X,y)
y10_predict = poly10_reg.predict(X)
#比较真实值和预测值的均方误差
display(mean_squared_error(y,y10_predict))
plt.scatter(x,y)
plt.plot(np.sort(x),y10_predict[np.argsort(x)],color='r')
plt.show()
- 多项式参数设置为degree=100
poly100_reg = PolynomialRegression(degree=100)
poly100_reg.fit(X,y)
y100_predict = poly100_reg.predict(X)
#比较真实值和预测值的均方误差
display(mean_squared_error(y,y100_predict))
plt.scatter(x,y)
plt.plot(np.sort(x),y100_predict[np.argsort(x)],color='r')
plt.show()

degree 越大,拟合的效果越好,因为样本点是一定的,我们总能找到一条曲线将所有的样本点拟合,也就是说让所有的样本点都落在这条曲线上,使得整体的均方误差为0; 曲线并不是所计算出的拟合曲线,只是原有的数据点对应的y的预测值连接出来的结果,而且有的地方没有数据点,因此连接的结果和原来的曲线不一样;而在未知的待预测数据的预测过程中,过量拟合训练集会造成泛化能力的降低,预测偏差增大。
这道题的欠拟合情况:
#欠拟合情况
clf = SGDRegressor(max_iter=500,tol=1e-2)
clf.fit(train_data,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data))
score_test = mean_squared_error(test_target,clf.predict(test_data))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)
过拟合情况
#过拟合情况
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(degree=5)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000,tol=1e-3)
clf.fit(train_data_poly,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data_poly))
score_test = mean_squared_error(test_target,clf.predict(test_data_poly))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)
#正常拟合情况
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(degree=3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000,tol=1e-3)
clf.fit(train_data_poly,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data_poly))
score_test = mean_squared_error(test_target,clf.predict(test_data_poly))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)

在正常拟合的情况下,模型在训练集和测试集上得到的 MSE值均为三种拟合情况下的较小值,此时模型很好地表达了数据关系
2、模型的泛化与正则化
1、泛化和正则化的概念
- 泛化:模型处理新样本的能力
- 正则化:给需要训练的目标函数加上一些规则(限制),目的是为了防止过拟合。在原始的损失函数后添加正则项,可以减小模型学习到的 θ \theta θ的大小,这样可以使模型的泛化能力更强。
-
- L1正则化使用L1范数,L1范数即向量元素绝对值之和。
∣ ∣ x ∣ ∣ 1 = ∑ i = 1 N ∣ x i ∣ ||x||_1=\sum_{i=1}^N|x_i| ∣∣x∣∣1=∑i=1N∣xi∣
- L1正则化使用L1范数,L1范数即向量元素绝对值之和。
-
- L2正则化使用L2范数【欧几里得范数】,即向量元素绝对值平方和再开方。
∣ ∣ x ∣ ∣ 2 = ( ∑ i = 1 n ∣ x i ∣ 2 ) 1 2 ||x||_2=(\sum_{i=1}^{n}|x_i|^2)^{\frac{1}{2}} ∣∣x∣∣2=(∑i=1n∣xi∣2)21
- L2正则化使用L2范数【欧几里得范数】,即向量元素绝对值平方和再开方。
加入正则化项后的代价函数(损失函数)为
J ( w ) = 1 2 ∑ j = 1 N { j i − w T σ ( x j ) } 2 + λ 2 ∑ j = 1 N ∣ w j ∣ q J(w)=\frac{1}{2}\sum_{j=1}^N{\{j_i-w^T\sigma(x_j)\}}^2+\frac{\lambda}{2}\sum_{j=1}^N|w_j|_q J(w)=21∑j=1N{ji−wTσ(xj)}2+2λ∑j=1N∣wj∣q
L2正则化
J = J 0 + α ∑ w 2 J = J_0+\alpha\sum w^2 J=J0+α∑w2
L1正则化
J = J 0 + α ∑ ∣ w 1 ∣ J = J_0+\alpha\sum |w^1| J=J0+α∑∣w1∣

2、岭回归和LASSO回归
- 岭回归:对参数空间进行L2范数正则化的线性模型
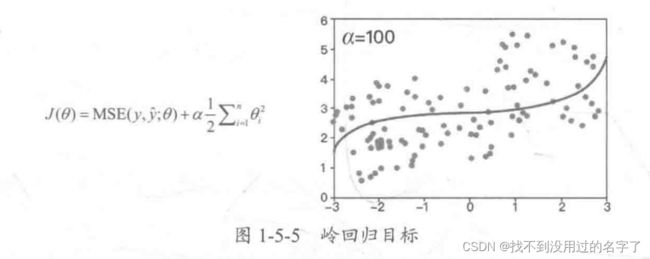
使用岭回归改进的多项式回归算法,随着 α \alpha α的改变,拟合曲线始终是曲线,直到最后变成一条几乎水平的直线;也就是说,在使用岭回归之后,多项式回归算法在模型变量前还是有系数的,因此很难得到一条斜的直线。 - LASSO回归:对参数空间进行L1范数正则化的线性模型

使用 LASSO 回归改进的多项式回归算法,随着 α \alpha α的改变,拟合曲线会很快变成一条斜的直线,最后慢慢变成一条几乎水平的直线,即模型更倾向于一条直线。
这道题的正则化:
#模型正则化L2
poly = PolynomialFeatures(degree=3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000,tol=1e-3,penalty='L2',alpha=0.0001)
clf.fit(train_data_poly,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data_poly))
score_test = mean_squared_error(test_target,clf.predict(test_data_poly))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)
#模型正则化L1
poly = PolynomialFeatures(degree=3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000,tol=1e-3,penalty='L1',alpha=0.0001)
clf.fit(train_data_poly,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data_poly))
score_test = mean_squared_error(test_target,clf.predict(test_data_poly))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)
#采用ElasticNet联合L1和L2范数加权正则化
poly = PolynomialFeatures(degree=3)
train_data_poly = poly.fit_transform(train_data)
test_data_poly = poly.transform(test_data)
clf = SGDRegressor(max_iter=1000,tol=1e-3,penalty='elasticnet',l1_ratio = 0.9,alpha=0.0001)
clf.fit(train_data_poly,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data_poly))
score_test = mean_squared_error(test_target,clf.predict(test_data_poly))
print('SGDRegressor train MSE: ',score_train)
print('SGDRegressor test MSE: ',score_test)
3、回归模型的评估指标和调用方法
- 平方绝对值误差(MAE):预测值和真实值之差的绝对值。【越小越好】
M A E = 1 n ∑ i = 1 n ∣ f i − y i ∣ = 1 n ∑ i = 1 n ∣ e i ∣ MAE = \frac{1}{n}\sum_{i=1}^{n}|f_i-y_i|=\frac{1}{n}\sum_{i=1}^n|e_i| MAE=n1∑i=1n∣fi−yi∣=n1∑i=1n∣ei∣
from sklearn .metrics import mean_absolute_error
mean_absolute_error(y_test,y_pred)
- 均方误差(MSE):参数估计值与参数真实值之差的平方期望值【越小越好】
M S E = 1 n ∑ i = 1 n ( o b s e r v e d i − p r e d i c t e d i ) 2 MSE = \frac{1}{n}\sum_{i=1}^n(observed_i-predicted_i)^2 MSE=n1∑i=1n(observedi−predictedi)2
from sklearn.metrics import mean_squared_error
mean_squared_error(y_test,y_pred)
- 均方根误差:是MSE的平方根
R M S E = M S E = 1 n ∑ i = 1 n w i ( y i − y i ^ ) 2 RMSE = \sqrt{MSE}=\sqrt{\frac{1}{n}\sum_{i=1}^nw_i(y_i-\hat{y_i})^2} RMSE=MSE=n1∑i=1nwi(yi−yi^)2
from sklearn.metrics import mean_squared_error
Pred_Error = mean_squared_error(y_test,y_pred)
Sqrt(Pred_Error)
- R平方值:回归模型在多大程度上解释了因变量的变化,或者说模型怼观测值的拟合程度如何。
R 2 ( y , y ^ ) = 1 − ∑ i = 0 n s a m p l e s − 1 ( y i − y i ^ ) 2 ∑ i = 0 n s a m p l e s − 1 ( y i − y i ‾ ) 2 R^2(y,\hat{y})=1-\frac{\sum_{i=0}^{n_{samples}-1}(y_i-\hat{y_i})^2}{\sum_{i=0}^{n_{samples}-1}(y_i-\overline{y_i})^2} R2(y,y^)=1−∑i=0nsamples−1(yi−yi)2∑i=0nsamples−1(yi−yi^)2
from sklearn.metrics import r2_score
r2_score(y_test,y_pred)
4、交叉验证
在某种意义下将原始数据进行分组, 一部分作为训练集,另 一部分作为验证集。首先用训练集对分类器进行训练,再利用验证集来测试训练得到的模型,以此来作为评价分类器的性能指标。
- 简单交叉验证
将原始数据随机分为两组。
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test= train_test_split(iris.data,iris.targe, test_size=.4, random_state=0)
- K折交叉验证
将原始数据分成K组(一般是均分) ,然后将每个子集数据分别做一次验证集,其余的 K-1组子集数据作为训练集,这样就会得到K个模型,将K个模型最终的验证集的分类准确率取平均值,作为K折交叉验证分类器的性能指标。通常设置K大于或等于3。
from sklearn.model_selection import Kfold
kf = KFold(n_splits=lO)
- 留一法交叉验证
每个训练集由除一个样本之外的其余样本组成,留下的一个样本组成检验集。这样 ,对于N个样本的数据集,可以组成N个不同的训练集和N个不同的检验集,因此会得到N个模型,用N个模型最终的验证集的分类准确率的平均数作为分类器的性能指标。
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut ()
- 留P法交叉验证
与留一法交叉验证类似,是从完整的数据集中删除p个样本,产生所有可能的训练集和检验集。
from sklearn.model_selection import LeavePOut
lpo = LeavePOut (p=5)
- 其他交叉验证分割方法
-
- (1)基于类标签,具有分层的交叉验证。【主要用于解决样本不平衡的问题。】
-
-
- StratifiedKFold K-Fold 的变种,会返回 stratified(分层)的折叠:在每个小集合中,各个类别的样例比例大致和完整数据集中的相同。
-
-
-
- StratifiedShuffleSplit是ShuffleSplit的一个变种,会返回直接的划分,比如创建一个划
分,但是划分中每个类的比例和完整数据集中的相同。
- StratifiedShuffleSplit是ShuffleSplit的一个变种,会返回直接的划分,比如创建一个划
-
-
- (2)用于分类数据的交叉验证**【进一步测试模型的泛化能力】**
-
-
- GroupKFold**【检测过拟合问题】**
K-Fold 变种,它确保同一个组在测试集和训练集中都不被表示。例如,如果数据是从不同的、有区别的组获得的,每个组又有多个样本,并且模型足够灵活,能高度从指定的特征中学习,则可能存在很好地拟合训练的组,但不能很好地预测不存在于训练组中的样本,而 GroupKFold 可以检测到这种过拟合的情况。
- GroupKFold**【检测过拟合问题】**
-
-
-
- LeaveOneGroupOut
根据用户提供的整数组的数组来区别不同组,以此来提供样本。这个组的信息可以用来编码任意域特定的预定义交叉验证折叠。每个训练集都是由除特定组别以外的所有样本构成的。
- LeaveOneGroupOut
-
-
-
- LeavePGroupsOut
类似于 LeaveOneGroupOut, 但是为了删除每个训练集/测试集与P组有关的样本。
- LeavePGroupsOut
-
-
-
- GroupShuffleSplit
ShuffleSplit 和 LeavePGroupsOut 的组合,生成一个随机划分分区的序列,其中为每个分组提供一个组子集。
- GroupShuffleSplit
-
-
- (3)时间序列分割
TimeSeriesSplit 也是 K-Fold 的一个变种,首先返回K折作为训练数据集,把 K+1折作为测试数据集。请注意,与标准的交叉验证方法不同,有关时间序列的样本切分必须保证时间上的顺序性,不能用未来的数据去验证现在数据的正确性,只能使用时间上之前一段的数据建模,而用后一段时间的数据来验证模型预测的效果,这也是时间序列数据在做模型验证划分数据时与其他常规数据切分的区别。
- (3)时间序列分割
这道题的交叉检验
#简单交叉检验
from sklearn.model_selection import train_test_split #切分数据
#切分数据,训练数据为 80%,验证数据为 20%
train_data, test_data, train_target, test_target =train_test_split(train, target, test_size=0.2, random_state=0)
clf= SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data))
score_test = mean_squared_error(test_target,clf.predict(test_data))
print ("SGDRegressor train MSE: ", score_train)
print("SGDRegressor test MSE: ", score_test)
#K折交叉检验
from sklearn.model_selection import KFold
kf = KFold(n_splits=5)
for k,(train_index, test_index) in enumerate(kf.split(train)):
train_data, test_data,train_target,test_target = train.values[train_index],train.values[test_index],target[train_index], target[test_index]
clf= SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data))
score_test = mean_squared_error(test_target,clf.predict(test_data))
print(k,"折", "SGDRegressor train MSE : ", score_train)
print(k,"折", "SGDRegressor test MSE " , score_test,'\n')
#留一法
from sklearn.model_selection import LeaveOneOut
loo = LeaveOneOut()
num = 100
for k,(train_index,test_index) in enumerate(loo.split(train)):
train_data,test_data,train_target,test_target = train.values[train_index],train.values[test_index],target[train_index],target[test_index]
clf = SGDRegressor(max_iter=1000,tol=1e-3)
clf.fit(train_data,train_target)
score_train = mean_squared_error(train_target,clf.predict(train_data))
score_test = mean_squared_error(test_target,clf.predict(test_data))
print(k,"个","SGDRegressor train MSE: ",score_train)
print(k,"个","SGDRegressor test MSE: ",score_test,'\n')
if k>= 9:
break
#留P法交叉验证
from sklearn.model_selection import LeavePOut
lpo = LeavePOut(p=10)
num = 100
for k, (train_index, test_index) in enumerate(lpo.split(train)):
train_data, test_data, train_target,test_target = train.values[train_index], train.values[test_index], target[train_index], target[test_index]
clf= SGDRegressor(max_iter=1000, tol=1e-3)
clf.fit(train_data, train_target)
score_train = mean_squared_error(train_target, clf.predict(train_data))
score_test = mean_squared_error(test_target, clf.predict(test_data))
print (k, "10个","SGDRegressor train MSE: ", score_train)
print (k, "10个","SGDRegressor test MSE: ", score_test,'\n')
if k >= 9:
break
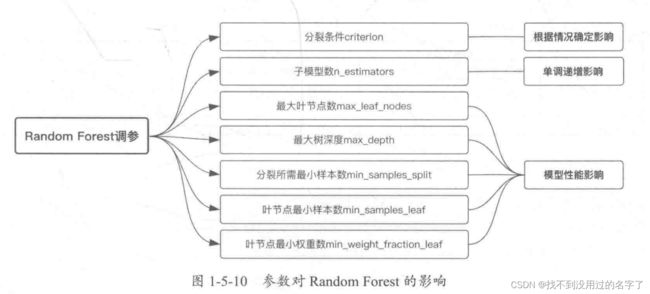
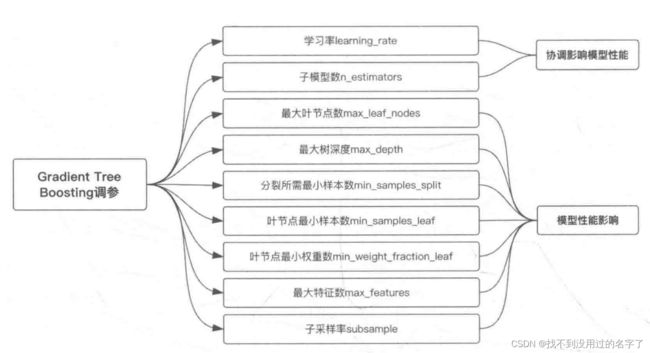
5、模型调参
1、参数影响
2、网格搜索
穷举搜索的调参手段。
from sklearn.datasets import load_iris
from sklearn.svm import SVC
from sklearn.model_selection import train_test_split
iris = load_iris()
X_train,X_test,y_train,y_test = train_test_split(iris.data,iris.target,random_state=0)
print("Size of training set:{} size of testing set:{}".format(X_train.shape[0],X_test.shape[0]))
#grid search start
best_score = 0
for gamma in [0.001,0.01,0.1,1,10,100] :
for C in [0.001,0.01,0.1,1,10,100]:
svm = SVC(gamma=gamma,C=C) #对于每种参数可能的组合 都进行一次训练
svm.fit(X_train,y_train)
score= svm.score(X_test,y_test)
if score > best_score : #找到表现最好的参数
best_score = score
best_parameters = {'gamma':gamma,'C':C}
### grid search end
print("Best score:{:.2f}".format(best_score))
print("Best parameters:{}".format(best_parameters))
#穷举网格搜索
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split #切分数据
#切分数据,训练数据为80%, 验证数据为 20%
train_data,test_data,train_target,test_target =train_test_split(train,target,test_size=0.2,random_state=0)
randomForestRegressor = RandomForestRegressor()
parameters = { 'n_estimators': [50,100,200] ,'max_depth': [1,2,3]}
clf= GridSearchCV(randomForestRegressor, parameters, cv=5)
clf.fit (train_data,train_target)
score_test = mean_squared_error(test_target,clf.predict(test_data))
print("RandornForestRegressor GridSearchCV tes MSE ", score_test)
sorted (clf.cv_results_.keys())
#随机参数优化
from sklearn.model_selection import RandomizedSearchCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split #切分数据
#切分数据, 练数据为 80% 验证数据为 20
train_data,test_data,train_target,test_target = train_test_split(train,target,test_size=0.2, random_state=0)
randomForestRegressor = RandomForestRegressor()
parameters = {
'n_estimators': [50 , 100 , 200 , 300] ,
'max_depth': [1 , 2 , 3 , 4 , 5 ]
}
clf= RandomizedSearchCV(randomForestRegressor, parameters, cv=5)
clf.fit(train_data,train_target)
score_test= mean_squared_error(test_target,clf.predict(test_data))
print ("RandomForestRegressor RandomizedSearchCV test MSE : ", score_test)
sorted (clf.cv_results_.keys ())
3、学习曲线
学习曲线是在训练集大小不同时,通过绘制模型训练集和交叉验证集上的准确率来观察模型在新数据上的表现,进而判断模型的方差或偏差是否过高,以及增大训练集是否可以减小过拟合。

这道题的学习曲线:
#学习曲线
import numpy as np
import matplotlib.pyplot as plt
from sklearn.linear_model import SGDRegressor
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import learning_curve
plt.figure(figsize=(18,10),dpi=150)
train_data2 = pd.read_csv('./zhengqi_train.txt', sep='\t')
test_data2 = pd.read_csv ('./zhengqi_test.txt', sep= '\t')
def plot_learning_curve(estimator,title,X,y,ylim=None,cv=None,n_jobs=1,train_sizes=np.linspace(.1,1.0,5)):
plt.figure()
plt.title(title)
if ylim is not None:
plt.ylim(*ylim)
plt.xlabel("Training examples")
train_sizes,train_scores,test_scores = learning_curve(estimator,X,y,cv=cv,n_jobs=n_jobs,train_sizes=train_sizes)
train_scores_mean = np.mean(train_scores,axis=1)
train_scores_std = np.std(train_scores,axis=1)
test_scores_mean = np.mean(test_scores,axis=1)
test_scores_std = np.std(test_scores,axis=1)
plt.grid()
plt.fill_between(train_sizes,train_scores_mean-train_scores_std,train_scores_mean+train_scores_std,alpha=0.1,color='r')
plt.fill_between(train_sizes,test_scores_mean-test_scores_std,test_scores_mean+test_scores_std,alpha=0.1,color='g')
plt.plot(train_sizes,train_scores_mean,'o-',color='r',label='Training score')
plt.plot(train_sizes,test_scores_mean,'o-',color='g',label='Cross-validation score')
plt.legend(loc='best')
return plt
X = train_data2[test_data2.columns].values
y = train_data2['target'].values
title = 'LinearRegression'
cv = ShuffleSplit(n_splits=100,test_size=0.2,random_state=0)
estimator = SGDRegressor()
plot_learning_curve(estimator,title,X,y,ylim=(0.7,1.01),cv=cv,n_jobs=-1)
4、验证曲线
和学习曲线不同,验证曲线的横轴为某个超参数的一系列值,由此比较不同参数设置下(而非不同训练集大小)模型的准确率。
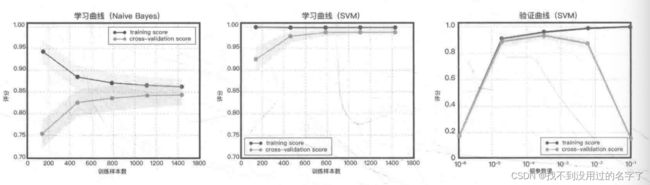
这道题的验证曲线
#验证曲线
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import SGDRegressor
from sklearn.model_selection import validation_curve
X = train_data2[test_data2.columns].values
y = train_data2['target'].values
param_range = [0.1,0.01,0.001,0.0001,0.00001,0.000001]
train_scores,test_scores = validation_curve(SGDRegressor(max_iter=1000,tol=1e-3,penalty='L1'),X,y,param_name='alpha',param_range=param_range,cv=10,scoring='r2',n_jobs=1)
train_scores_mean = np.mean(train_scores,axis=1)
train_scores_std = np.std(train_scores,axis=1)
test_scores_mean = np.mean(test_scores,axis=1)
test_scores_std = np.std(test_scores,axis=1)
plt.title("Validation Curve with SGDRegressor")
plt.xlabel('alpha')
plt.ylabel('Score')
plt.ylim(0.0,1.1)
plt.semilogx(param_range, train_scores_mean, label="Training score", color= "r")
plt.fill_between(param_range, train_scores_mean - train_scores_std, train_scores_mean+ train_scores_std, alpha=0.2, color="r")
plt.semilogx(param_range,test_scores_mean, label="Cross-validation score", color="g")
plt.fill_between(param_range, test_scores_mean - test_scores_std,test_scores_mean+ test_scores_std, alpha=0.2, color="g")
plt.legend(loc="best")
plt.show ()
8、特征优化
特征优化方法
- 合成特征
指不在输入特征之列,而是从一个或多个输入特征衍生而来的特征。通过标准化或缩放单独创建的特征不属于合成特征。合成特征包括以下类型: -
- (1)将 个特征与其本身或其他特征相乘(称为特征组合)。
-
- (2) 两个特征相除
-
- (3) 对连续特征进行分桶(分箱),以分为多个区间分箱
- 特征的简单变换
-
- 数值特征的变换和组合
-
-
- (1)多项式特征 polynomial feature)
-
-
-
- (2) 比例特征 (ratio feature): X 1 / X 2 X1/X2 X1/X2
-
-
-
- (3) 绝对值 (absolute value)
-
-
-
- (4) max (X1,X2), min (X1,X2), X1 or X2
-
-
- 类别特征与数值特征的组合
用N1和N2表示数值特征,用C1和C2表示类别特征,利用 Pandas的groupby 操作可以创造出以下几种有意义的新特征(其中,C2 还可以是离散化了的N1)
- 类别特征与数值特征的组合
-
-
- (1)中位数: median(N1) _by(C1)
-
-
-
- (2)算术平均数: mean(N1) _ by(C1)
-
-
-
- (3)众数:mode(N1) _by(C1)
-
-
-
- (4)最小值:min(N1) _by(C1)
-
-
-
- (5)最大值:max(N1)_by(C1)
-
-
-
- (6)标准差:std(N1) _by(C1)
-
-
-
- (7)方差:var(N1) _by(C1)
-
-
-
- (8)频数:freq (C2) _ (C1)【freq(C1) 不需要进行 groupby 操作也有意义】
-
- 用决策树创造新特征
在决策树系列(单棵决策树、GBDT、随机森林)的算法中,由于每一个样本都会被映射到决策树的一片叶子上,因此我们可以把样本经过每一棵决策树映射后的 index (自然数)或 one-hot-vector (哑编码得到的稀疏矢量)作为一项新的特征加入模型中。
具体实现可以采用 apply() 方法和 decision_path()方法,其在 sklearn 和 xgboost 中都可以用。 - 特征组合
-
- 组合独热矢量
-
- 使用分桶特征列训练模型
以一定方式将连续型数值特征划分到不同的桶(箱)中,可以理解为是对
连续型特征的一种离散化处理方式。
- 使用分桶特征列训练模型
epsilon = 1e-5
func_dict = {
'add':lambda x,y:x+y,
'min':lambda x,y:x-y,
'div':lambda x,y:x/(y+epsilon),
'multi':lambda x,y:x*y
}
def auto_features_make(train_data,test_data,func_dict,col_list):
train_data,test_data = train_data.copy(),test_data.copy()
for col_i in col_list:
for col_j in col_list:
for func_name,func in func_dict.items():
for data in [trian_data,test_data]:
func_features = func(data[col_i],data[col_j])
col_func_features = '-'.join([col_i,func_name,col_j])
data[col_func_features] = func_features
return train_data,test_data
9、模型融合
1、模型优化
(1)如何判断模型的优劣及优化模型的性能呢?
- 研究模型学习曲线,判断模型是否过拟合或者欠拟合并做出相应的调整;
- 对模型权重参数进行分析,对于权重绝对值高或低的特征,可以进行更细化的工作,也可以进行特征组合;
- 进行 Bad-Case 分析,针对错误的例子确定是否还有地方可以修改挖掘;
- 进行模型融合。
(2)模型融合提升技术
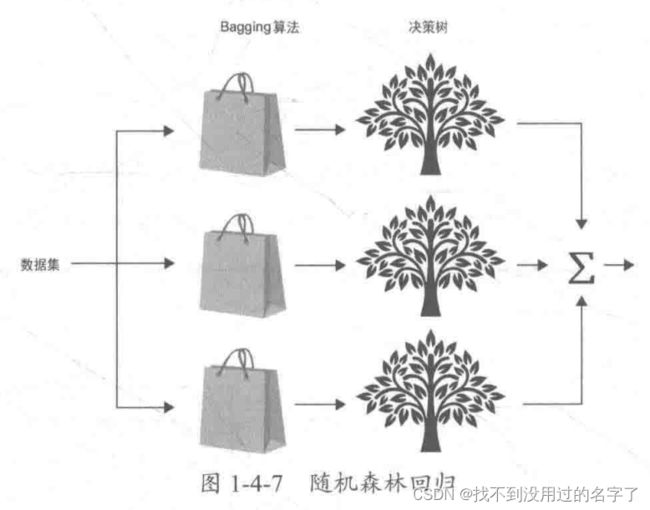
- 个体学习器间不存在强依赖关系可同时生成的并行化方法,代表是 Bagging 方法和随机森林。
-
- 随机森林是对 Bagging 方法的改进,其改进之处有两点:基本学习器限定为决策树;除了在 Bagging 的样本上加上扰动,在属性上也加上扰动,相当于在决策树学习的过程中引入了随机属性选择。对基决策树的每个结点,先从该结点的属性集合中随机选择一个包含k个属性的子集 ,然后从这个子集中选择一个最优属性用于划分。
- 个体学习器间存在强依赖关系必须串行生成的序列化方法,代表是 Boosting 方法。
Boosting 方法的训练过程为阶梯状,即基模型按次序一一进行训练(实现上可以做到并行),基模型的训练集按照某种策略每次进行一定的转换,然后对所有基模型预测的结果进行线性综合,产生最终的预测结果。
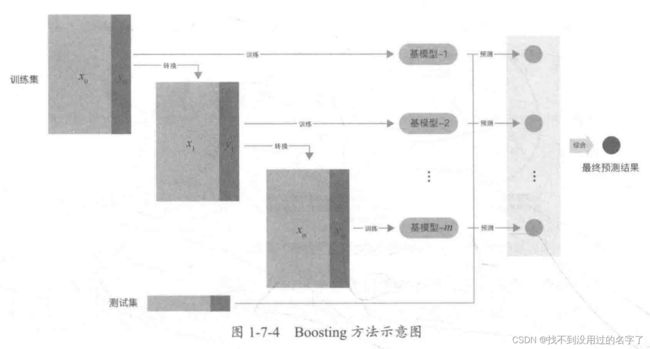
-
- AdaBoost 算法:是加法模型、损失函数为指数函数、学习算法为前向分布算法时的二分类算法。
-
- 提升树:是加法模型、学习算法为前向分布算法时的算法,基本学习器限定为决策树。对于二分类问题,损失函数为指数函数,就是把 AdaBoost 算法中的基本学习器限定为二叉决策树;对于回归问题,损失函数为平方误差,此时拟合的是当前模型的残差。
-
- 梯度提升树:是对提升树算法的改进。提升树算法只适合于误差函数为指数函数和平方误差,而对于一般的损失函数,梯度提升树算法可以将损失函数的负梯度在当前模型的值作为残差的近似值。
(3)预测结果融合策略
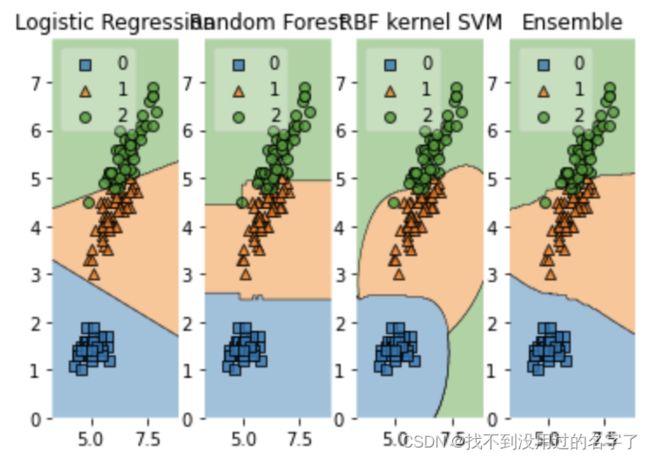
- Voting
Voting (投票机制)分为软投票和硬投票两种,其原理采用少数服从多数的思想,此方法可用于解决分类问题。
- 硬投票:对多个模型直接进行投票,最终投票数最多的类为最终被预测的类。
- 软投票:和硬投票原理相同,其增加了设置权重的功能,可以为不同模型设置不同权重,进而区别模型不同的重要度。
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import itertools
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from mlxtend.classifier import EnsembleVoteClassifier
from mlxtend.data import iris_data
from mlxtend.plotting import plot_decision_regions
clf1 = LogisticRegression(random_state=0,solver='lbfgs',multi_class='auto')
clf2 = RandomForestClassifier(random_state=0,n_estimators=100)
clf3 = SVC(random_state=0,probability=True,gamma='auto')
eclf = EnsembleVoteClassifier(clfs=[clf1,clf2,clf3],weights=[2,1,1],voting='soft')
X,y = iris_data()
X = X[:,[0,2]]
gs = gridspec.GridSpec(1,4)
fig = plt.figure(figsize=(16,4))
for clf,lab,grd in zip([clf1,clf2,clf3,eclf],['Logistic Regression','Random Forest','RBF kernel SVM','Ensemble'],itertools.product([0,1],repeat=2)):
clf.fit(X,y)
ax = plt.subplot(gs[0,grd[0]*2+grd[1]])
fig = plot_decision_regions(X=X,y=y,clf=clf,legend=2)
plt.title(lab)
-
- Averaging 的原理是将模型结果的平均值作为最终的预测值,也可以使用加权平均的方法。但其也存在问题:如果不同回归方法预测结果的波动幅度相差比较大,那么波动小的回归结果在融合时起的作用就比较小。
-
- Ranking 的思想和 Averaging 的一致。因为上述平均法存在一定的问题,所以这里采用了把排名平均的方法。如果有权重,则求出n个模型权重比排名之和,即为最后的结果。
- Blending
Blending 把原始的训练集先分成两部分 ,如 70%的数据作为新的训练集,剩下 30%的数据作为测试集。
在第一层中,我们用 70%的数据训练多个模型,然后去预测剩余 30%数据的 label。 在第二层中,直接用 30%的数据在第一层预测的结果作为新特征继续训练即可。
Blending的优点:Blending 比 Stacking 简单(不用进行 k 次交叉验证来获得 stacker feature), 避开了一些信息泄露问题,因为 generlizers 和 stacker 使用了不一样的数据。
Blending 的缺点:
(I) 使用了很少的数据 (第二阶段的 blender 只使用了训练集 10%的数据量)
( 2) blender 可能会过拟合。
说明:对于实践中的结果而言, Stacking Blending 的效果差不多。 - Stacking
Stacking 是一种分层模型集成框架。以两层为例:第一层由多个基学习器组成,其输入为原始训练集;第二层的模型则是以第一层基学习器的输出作为训练集进行训练,从而得到完整的 Stacking 模型。Stacking 层模型都使用了全部的训练集数据。
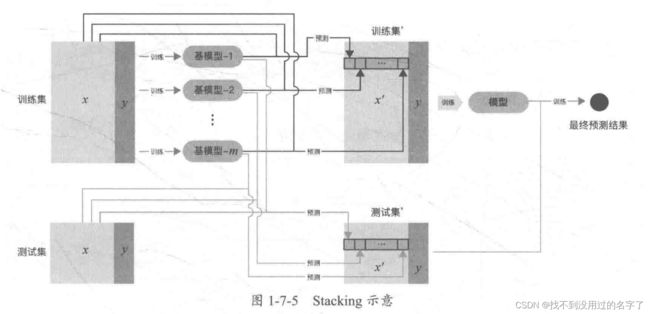
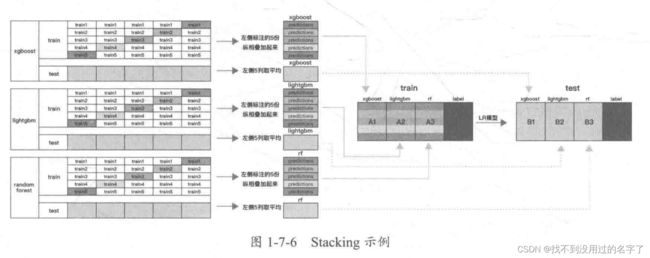
2、单一模型预测效果
- 岭回归
model='Ridge'
opt_models[model]=Ridge()
alph_range = np.arange(0.25,6,0.25)
param_grid = {'alpha':alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model], param_grid=param_grid,splits=splits, repeats=repeats)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range,abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits*repeats))
plt.xlabel('alpha')
plt.ylabel('score')
(1)真实值(横轴:y) 与模型预测值(竖轴:y_pred)的散点图,图形上方显示了相关性数值,其越接近1越好。对于岭回归模型,相关性数值为 0.947, 预测值与真实值比较一致。
(2) 其为在交叉验证训练模型时,真实值(横轴:y) 与栈型预测值和真实值的残 (竖轴: y - y_pred)的散点图,图形上方显示了方差,其越小说明模型越稳定。可以看到,对于岭回归模型,在真实值 y=-3 附近的预测值有较大的偏差,同时,方差为 0.319, 较为稳定。
(3) 图是由模型预测值和真实值的残差(横轴: z = (resid - mean_resid) / std_resid)
落在按 z 轴划分区间的频率(竖轴:频数)所画的直方图,图形上方显示了预测值与真实值的残差大于三倍标准差的数,其越小越好,越大说明预测中有些样本的偏差很大。对于岭回归模型,预测值与真实值的残差大于三倍标准差的数为 5 个,模型对偏差大的数据有较好的包容性。
(4) 岭回归模型的参数(横轴 :alpha) 与模型的评价指标 MSE (竖轴: score)的误差棒图。
2. Lasso 回归
model ='Lasso'
opt_models[model] = Lasso()
alph_range = np.arange(1e-4,1e-3,4e-5)
param_grid = {'alpha': alph_range}
opt_models[model],cv_score,grid_results = train_model(opt_models[model],param_grid=param_grid,splits=splits,repeats=repeats)
cv_score.name= model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(alph_range,abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqrt(splits * repeats))
plt.xlabel ('alpha')
plt.ylabel ('score')
- ElasticNet 回归
model='ElasticNet'
opt_models[model] = ElasticNet()
param_grid = {'alpha': np.arange(1e-4, 1e-3, 1e-4),'ll_ratio': np.arange(0.1,1.0,0.1), 'max iter': [100000]}
opt_models[model], cv_score, grid_results = train_model(opt_models[model],param_grid=param_grid,splits=splits,repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
- SVR 回归
model='LinearSVR'
opt_rnodels[model] = LinearSVR()
crange = np.arange(0.1,1.0,0.1)
param_grid = {'C':crange,'max_iter':[1000]}
opt_models[model],cv_score,grid_results = train_model(opt_models[model],param_grid=param_grid,splits=splits,repeats=repeats)
cv_score.name= model
score_models = score_models.append(cv_score)
plt.figure()
plt.errorbar(crange,abs(grid_results['mean_test_score']),abs(grid_results['std_test_score'])/np.sqr(splits * repeats))
plt.xlabel('C')
plt.ylabel('score')
- K近邻
model='KNeighbors'
opt_models[model] = KNeighborsRegressor()
param_grid = {'n_neighbors': np.arange(3,11,1)}
opt_models[model], cv_score, grid_results = train_model(opt_models[model],param_grid=param_grid,splits=splits,repeats=1)
cv_score.name = model
score_models = score_models.append(cv_score)
plt.figure ()
plt.errorbar(np.arange(3, 11, 1),
abs(grid_results['mean_test_score']),
abs(grid_results['mean_test_score']) /np.sqrt(splits * 1))
plt.xlabel('n_neighbors')
plt.ylabel('score')
3、多模型融合方法
1、Boosting方法
from sklearn.model_selection import KFold
import pandas as pd
import numpy as np
from scipy import sparse
import xgboost
import lightgbm
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import AdaBoostRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
def stacking_reg(clf, train_x , train_y , test_x , clf_name , kf,
label_split=None):
train= np.zeros((train_x.shape[0],1))
test = np.zeros((test_x.shape[0],1))
test_pre = np.empty((folds, test_x.shape[O], 1))
cv_scores= []
for i,(train_index,test_index) in enumerate(kf.split(train_x, label split)):
tr_x = train_x[train_index]
tr_y = train_y[train_index]
te_x= train_x[test_index]
te_y = train_y[test_index]
if clf_name in ["rf","ada", "gb", "et", "lr", "lsvc", "knn"]:
clf.fit (tr_x , tr_y)
pre= clf.predict(te_x).reshape (-1,1)
train[test_index] = pre
test_pre[i,:] = clf.predict(test_x).reshape (-1,1)
cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["xgb"]:
train_matrix = clf.DMatrix(tr_x, label=tr_y,missing=-1)
test_matrix = clf.DMatrix(te_x, label=te_y, missing=-1)
z = clf.DMatrix (test_x,label=te_y,missing=-1)
params = { 'booster' : 'gbtree',
'eval_metric' : 'rmse',
'gamma' : 1 ,
'min_child_weight': 1.5,
'max_ ep': 5,
'lambda' : 10 ,
'subsample': 0.7 ,
'colsaMple_bytree' : 0.7,
'colsample_bylevel': 0.7,
'eta' : 0.03 ,
'tree_ method':'exact',
'seed': 2017 ,
'nthread': 12
}
num_round= 10000
early_stopping_rounds = 100
watchlist =[(train_matrix,'train'), (test_matrix ,'eval')]
if test_matrix:
model = clf.train(params, train_matrix,num_boost_round=num_round, evals=watchlist, early_stopping_rounds=early_stopping_rounds)
pre = model.predict(test_matrix, ntree_limit=model_best_ntree_limit).reshape(-1 , 1)
train[test_index] = pre
test_pre[i, :] =model.predict(z, ntree_limit=model.best_ntree_limit).reshape(-1, 1)cv_scores.append(mean_squared_error(te_y, pre))
elif clf_name in ["lgb"]:
train_matrix = clf.Dataset(tr_x, label=tr_y)
test_matrix = clf.Dataset(te_x, label=te_y)
params = {
'boosting_ type':'gbdt',
'objective':'regression_12',
'metric':'mse',
'min_child_weight': 1.5,
'num leaves': 2**5,
'lambda 12': 10,
'subsample': 0. 7,
'colsample_bytree': 0. 7,
'colsample_bylevel': 0.7,
'learning_rate': 0. 03,
'tree_ method':'exact',
'seed': 2017,
'nth read': 12,
'silent': True,
}
nuro_round = 10000
early_stopping_rounds = 100
if test_matrix:
model = clf.train (
params,
train_matrix,
num_round,
valid_sets=test_matrix,
early_stopping_rounds=early_stopping_rounds)
pre = model.predict(te_x,num_iteration=model.best_iteration).reshape (-1, 1)
train[test_index]= pre
test_pre[i, :] = model.predict(test_x, num_iteration=model.best_iteration).reshape(-1, 1)
cv_scores.append(mean_squared_error(te_y, pre))
else:
raise IOError ("Please add new clf.")
print("%s now score is:" % clf_name, cv_scores)
test(:]= test_pre.mean(axis=O)
print("%s_score_list:" % clf_name, cv_scores)
print("%s_score_mean: "% clf_name, np.mean{cv_scores))
return train.reshape(-1, 1), test.reshape(-1, 1)
2、Stacking方法
def rf_reg(x_train, y_train, x_valid, kf, label_split=None):
randomforest = RandomForestRegressor(n_estimators=600,max_depth=20, n_jobs=-1, random_state=2017,max_features="auto", verbose=1)
rf_train, rf_test = stacking_reg(randomforest, x_train, y_train , x_valid, "rf", kf, label_ split=label_split)
return rf_train, rf_test, "rf_reg"
def ada_reg(x_train, y_train, x_valid, kf, label_split=None) :
adaboost = AdaBoostRegressor(n_estimators=30,random_state=2017, learning_rate=0.01)
ada_train, ada_test= stacking_reg(adaboost,x_train, y_train, x_valid, "ada", kf, label split= label_split)
return ada_train,ada_test, "ada_reg"
def gb_reg(x_train, y_train, x_valid, kf, label_split=None):
gbdt = GradientBoostingRegressor(learning_rate=0.04,n_estimators=100, subsample=0.8, random_state=2017, max_depth=5,verbose=1)
gbdt_train, gbdt_test= stacking_reg(gbdt,x_train, y_train,x_valid,"gb",kf,label_split=label_split)
return gbdt_trian,gbdt_test,"gb_reg"
def et_reg(x_train, y_train,x_valid,kf,label_split=None):
extratree = ExtraTreesRegressor(n_estimators=600,max_depth=35,max_features="auto", n_jobs=-1 , random_state=2017,verbose=1)
et_train, et_test= stacking_reg(extratree,x_train, y_train, x_valid, "et", kf, label_split=label_split)
return et_train, et_test ,"et_reg"
def lr_reg(x_train , y_train,x_valid, kf, label_split=None):
lr_reg = LinearRegression(n_jobs=- 1)
lr_train, lr_test = stacking_reg(lr_reg,x_train,y_train, x_valid, "lr", kf, label_split=label_split)
return lr_train,lr_test,"lr_reg"
def xgb_reg(x_train,y_train,x_valid, kf, label_split=None):
xgb_train , xgb_test = stacking_reg(xgboost,x_train, y_train, x_valid, "xgb", kf, label_split= label_split)
return xgb_train, xgb_test,"xgb_reg"
def lgb_reg(x_train,y_train,x_valid, kf, label_split=None):
lgb_train, lgb_test= stacking_reg(lightgbm,x_train,y_train, x_valid, "lgb", kf, label_split=label_split)
return lgb_train, lgb_test, "lgb_reg"
4、定义模型融合 Stacking 预测函数
对模型融合 Stacking 的预测函数进行定义
def stacking_pred(x_train,y_train, x_valid, kf,clf_list,label_split=None, clf_fin="lgb", if_concat_origin=True):
for k, clf_list in enumerate(clf_list):
clf_list = [clf_list]
column_list = []
train_data_list = []
test_data_list= []
for clf in clf_list:
train_data, test_data, clf_name= clf(x_train,y_train, x_valid, kf, label_split=label_split)
train_data_list.append(train_data)
test_data_list.append(test_data)
column_list.append("clf_%s" % (clf_name))
train= np.concatenate(train_data_list, axis=1)
test= np.concatenate(test_data_list, axis=1)
if if_concat_origin:
train= np.concatenate([x_train, train], axis=1)
test= np.concatenate([x_valid, test], axis=1)
print(x_train.shape)
print(train.shape)
print(clf_name)
print(clf_name in ["lgb"])
if clf_fin in [ "rf", "ada", "gb", "et", "lr", "lsvc", "knn"]:
if clf_fin in ["rf"]:
clf= RandomForestRegressor(n_estimators=600,max_depth=20, n_jobs=-1, random_state=2017, max_features="auto" , verbose=1)
elif clf_fin in ["ada"] :
clf = AdaBoostRegressor (n_estimators=30,random_stat e=2017,learning_rate=0.01)
elif clf_fin in ["gb"]:
clf= GradientBoostingRegressor(learning_rate=0.04, n_estimators=100,subsample=0.8, random_state=2017 ,max_depth=5,verbose=1)
elif clf_fin in ["et"] :
clf= ExtraTreesRegressor(n_estimators=600,max_depth=35 , max_features="auto",n_jobs=-1 , random_state=2017,verbose=1)
elif clf_fin in [ "lr"] :
clf= LinearRegression(n_jobs=-1)
clf.fit(train, y_train)
pre= clf.predict(test).reshape(-1,1)
return pred _
elif clf_fin in ["xgb"]:
clf = xgboost
train_matrix = clf. DMatrix(train, label=y_train,missing=-1)
test_matrix = clf.DMatrix(train , label=y_train,missing=-1)
params = {
' booster':'gbtree',
' eval _metric ':'rmse',
'gamma ': 1 ,
'min_child_weight' : 1.5,
'max_depth' : 5,
'lambda': 10,
'subsample': 0.7,
'colsample_bytree': 0.7 ,
'colsample_bylevel': 0.7 ,
'eta': 0.03 ,
'tree_method' :'exact',
'seed': 2017,
'nthread': 12
}
num_round = 10000
early_stopping_rounds = 100
watchlist = [(train_matrix, 'train'), (test_matrix ,' eval')]
model = clf. train(params, train_matrix, num_boost_round=num_round, evals=watchlist, early_stopping_rounds=early_stopping_rounds)
pre = model.predict(test, ntree_limit=model.best_ntree_limit).reshape(-1, 1)
return pre
elif clf_fin in ["lgb"]:
print(clf_name)
clf= lightgbm
train_matrix = clf.Dataset(train, label=y_train)
test_matrix = clf.Dataset(train, label=y_train)
params = {
'boosting_type ':' gbd ',
'objective':'regression_l2',
'metric':'mse',
'min_child_weight' : 1. 5,
'num_leaves' : 2**5 ,
'lambda_12' : 10,
'subsample': 0. 7,
'colsample_bytree': 0.7,
'colsample_bylevel': 0.7,
'learning_rate': 0. 03,
'tree_method' :' exact ',
'seed': 2017,
'nthread': 12,
'silent': True,
}
num_round = 10000
early_stopping_rounds = 100
model = clf.train(params,train_matrix, num_round, valid_sets =test_matrix,early_stopping_rounds=early_stopping_rounds)
print (' pred ' )
pre = model.predict(test,num_iteration=model.best_itera ion.rehape(- 1 , 1 )
print (pre)
return pre
5、模型验证
- 加载数据
with open(". /zhengqi_train.txt") as fr:
data_train=pd.read_table(fr, sep="\t")
with open( ". /zhengqi_test.txt ") as fr_test:
data_test = pd.read_table(fr_test, sep="\t")
- 5折交叉验证
from sklearn.model_selection import StratifiedKFold, KFold
folds= 5
seed= 1
kf = KFold(n_splits=5 , shuffle=True , random_state=0)
- 训练集数据和测试集数据
x_train = data_train[data_test.columns].values
x_valid = data_test[data_test.columns].values
y_train = data_train['target'].values
- 使用 lr_reg 和 lgb_reg 进行融合预测
pred = stacking_pred(x_train, y_train , x_valid, kf, clf_list, label_split=None , clf_fin="lgb", if_concat_origin=True)