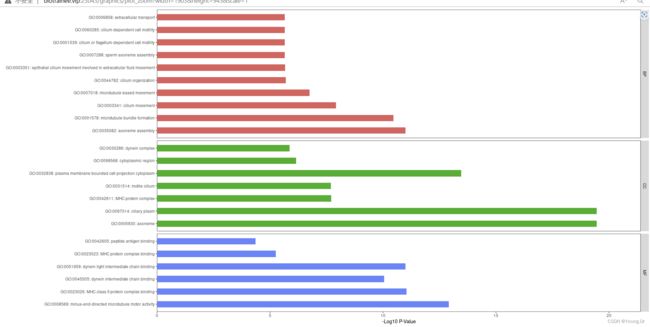
- bj某dongfang医院js 反混淆 分析结果
yyds970108
javascript开发语言ecmascript算法
没啥特别的手段,就是反混淆,动态调试+替换constCryptoJS=require('crypto-js');constforge=require('node-forge');constaxios=require('axios');const{v4:uuidv4}=require('uuid');constqs=require('querystring');functioncreateRando
- 【Python LeetCode 专题】热题 100,重在思路
一杯水果茶!
人生苦短我用Pythonpythonleetcode
哈希1.两数之和49.字母异位词分组128.最长连续序列双指针283.移动零11.盛最多水的容器15.三数之和42.接雨水滑动窗口3.无重复字符的最长子串438.找到字符串中所有字母异位词子串560.和为K的子数组239.滑动窗口最大值普通数组53.最大子数组和56.合并区间189.轮转数组238.除自身以外数组的乘积矩阵73.矩阵置零链表160.相交链表206.反转链表234.回文链表141.环
- JAVA刷题记录: 专题十五 BFS解决FloodFill算法
用屁屁笑
宽度优先算法
733.图像渲染-力扣(LeetCode)classSolution{int[]dx={0,0,-1,1};int[]dy={1,-1,0,0};publicint[][]floodFill(int[][]image,intsr,intsc,intcolor){intprev=image[sr][sc];if(color==prev)returnimage;Queueq=newLinkedList
- 自己开发I2C Bootloader -上位机开发篇
EE工程师
嵌入式系统pythonstm32单片机
上位机脚本开发 在芯片原厂大部分工程师选择的脚本语言依然是Python,Python有哪些开发优势这里就不再讨论了,这里我们只陈述一下上位机的开发环境,作者的开发环境是VSCode+Anaconda。脚本内容也没有什么好说的,一看就懂,比较简单。唯一值得提醒的是本项目的上位机开发需要多注意*Write_DataBytes_To_Serial_Port(self,DataBytes):*函数的实现
- ZooKeeper学习专栏(一):分布式协调的核心基石
快乐肚皮
Zookeeper分布式zookeeper学习
文章目录前言一、ZooKeeper是什么?二、为什么需要分布式协调服务?三、核心数据模型:ZNode3.1树形命名空间:分布式世界的文件系统3.2ZNode类型3.3ZNode数据结构:数据+元数据的完美融合Stat核心字段解析3.4ZNode操作3.5ZNode设计哲学3.6实战代码总结前言在分布式系统蓬勃发展的时代,我们享受着高并发、高可用的服务,却鲜少思考背后的协调艺术。当数百个服务节点部署
- 14.优化算法之BFS解决FloodFill算法1
muyierfly
算法题算法宽度优先深度优先
0.FloodFill简介dfs:深度优先遍历(红色)bfs:宽度优先遍历1.图像渲染算法原理classSolution{int[]dx={0,0,1,-1};int[]dy={1,-1,0,0};publicint[][]floodFill(int[][]image,intsr,intsc,intcolor){intprev=image[sr][sc];//统计刚开始的颜⾊if(prev==co
- BFS 解决 FloodFill 算法(C++)
lim 鹏哥
刷题算法宽度优先c++
文章目录前言一、概念二、岛屿数量1.题目链接2.算法原理3.代码编写三、被围绕的区域1.题目链接2.算法原理3.代码编写总结前言一、概念BFS就是广度优先遍历,也就是层序遍历。FloodFill是指在数组中找出性质相同的连通块,并根据题目进行操作。二、岛屿数量1.题目链接200.岛屿数量2.算法原理遍历整个矩阵,每找到一块陆地,记录一次。我们怎末知道我们是否已经遍历过这个地方了呢??方法1:如果遍
- BFS-FloodFill 算法 解决最短路问题 多源 解决拓扑排序
penguin_bark
#BFS算法宽度优先leetcode
文章目录一、FloodFill算法[733.图像渲染](https://leetcode.cn/problems/flood-fill/description/)2.思路3.代码[200.岛屿数量](https://leetcode.cn/problems/number-of-islands/description/)2.思路3.代码[LCR105.岛屿的最大面积](https://leetcod
- 解决Flutter运行android提示Deprecated imperative apply of Flutter‘s Gradle plugins
旺仔大牛
gradleflutterandroidgraldebuildscriptdependenciesprepositories
文章目录出现场景解决方案编辑android/settings.gradle编辑android/build.gradle重新定义库变量编辑android/app/build.gradle删除fluttetRoot和plugin字段添加plugins块修改dependencies出现场景ado@adodeMacBook-Airapp_demo%flutterrun--profileLaunchingl
- Vue.config全局配置
辉夜真是太可爱啦
1.取消Vue所有的日志与警告。Vue.config.silent=true;2.Vue自定义键名Vue.config.keyCodes={v:86,f1:112,//camelCase不可用mediaPlayPause:179,//取而代之的是kebab-case且用双引号括起来"media-play-pause":179,up:[38,87]}3.设置为false以阻止vue在启动时生成生产提
- Spring AI-15.Spring AI API
程序员勇哥
人工智能(AI)Java全套教程SpringAI人工智能springjavaSpringAI
SpringAI-15.SpringAIAPISpringAIAPI涵盖了广泛的功能。每个主要功能都在其专门的部分中详细介绍。以下是可用的关键功能概述:简介SpringAIAPI提供跨AI供应商的可移植模型API,适用于聊天、文本转图像、音频转录、文本转语音和嵌入模型。支持同步和流式API选项,同时也支持访问特定于模型的功能。AI模型API支持的模型类型:聊天模型(ChatModel):处理对话交
- 今日欧美圈:全英音乐奖获奖名单速递,哈卷情人节遭持刀抢劫
胡萝卜音乐
今天凌晨进行的2020年全英音乐奖完整获奖名单如下:年度专辑:Dave-"PSYCHODRAMA"年度单曲:LewisCapaldi-"SomeoneYouLoved"最佳男歌手:Stormzy最佳女歌手:Mabel最佳组合:Foals最佳新人:LewisCapaldi最佳国际男歌手:Tyler,TheCreator最佳国际女歌手:BillieEilish潜力新星:Celeste英媒报道Harry
- 【python】图片批量压缩脚本
横桥码农
pythonpython
#-*-coding:utf-8-*-'''图片批量压缩脚本将脚本放入待压缩文件夹下,并运行自动生成压缩文件夹compress'''fromPILimportImageimportosimportsysimportiosys.stdout=io.TextIOWrapper(sys.stdout.buffer,encoding='utf-8')defcompress_image(input_imag
- 001双双-文案课第七次作业
双双执行力财富流教练
作业要求:竞品分析做一个手机的竞品分析至于选择哪两款产品出于什么目的进行分析,需要按照韩老白老师今天讲的四个步骤来对比机型:iPhoneXvs坚果R1iPhoneXvs坚果R1参考资料:iPhoneX参数:http://product.pconline.com.cn/mobile/apple/1048848_detail.html坚果R1参数:http://product.pconline.com
- 关于事务的传播方式
ruan114514
java数据库开发语言mysql
传播行为定义了当一个事务方法被另一个事务方法调用时,事务应该如何进行。它决定了新方法是加入已有的“事务组”,还是自己单开一个“新项目组”,或者干脆不参与“项目组”(事务)。核心传播行为详解与示例:假设我们有两个服务方法:ServiceA.methodA()和ServiceB.methodB()。methodA内部调用了methodB。1.REQUIRED(默认)-“一起干”或“开新组”含义:如果当
- AirPlay认证是什么?AirPlay认证流程有哪些(ai)
Microtest_CS
AirPlay认证
在当今日益数字化的世界中,无线连接技术已成为我们日常生活中不可或缺的一部分。其中,AirPlay作为苹果公司推出的一种无线媒体播放技术,为用户提供了将音频、视频和照片等内容从iOS设备、Mac电脑等发送到AppleTV、HomePod或其他兼容设备的便捷方式。然而,为了确保用户能够获得最佳体验,苹果公司对于支持AirPlay的设备或软件有着严格的认证流程,这就是所谓的AirPlay认证。一、Air
- 华为HiCar认证
Microtest_CS
华为服务器运维
华为HiCar认证是针对车载智能互联系统的一项认证,旨在确保设备和应用能够与华为HiCar系统无缝连接和交互,从而为用户提供智慧出行体验。通过认证,设备和应用能够获得华为的官方认可,提升产品竞争力和市场认可度。华为HiCar认证项目:1.功能兼容性测试:验证设备和服务是否能与HiCar系统正常连接,以及各项功能是否能够在车载环境中正常运行;2.性能测试:对设备和服务进行性能测试,包括响应时间、数据
- 河道污染难溯源?3步搭建陌讯实时目标检测系统
2501_92472966
目标检测人工智能计算机视觉算法视觉检测
开篇痛点「凌晨3点水泵房渗漏报警,运维人员冒雨排查却是一场误判」——这是某水务企业技术总监向我吐槽的真实案例。在智慧水务场景中,传统视觉算法面临三大死穴:水体反光干扰、微小目标漏检、边缘设备算力受限。尤其当暴雨导致水体浑浊时,OpenCV边缘检测的误报率可达35%以上。技术解析:陌讯多模态融合架构为解决复杂环境泛化问题,陌讯视觉算法提出FMT-Net(FusionMultimodalTransfo
- LeetCode 72. 编辑距离(Edit Distance)| 动态规划详解
72.编辑距离题目描述给你两个单词word1和word2,请计算将word1转换为word2所需的最少操作数。你可以对一个单词进行以下三种操作:插入一个字符删除一个字符替换一个字符✅示例输入:word1="horse",word2="ros"输出:3解释:horse->rorse(替换h为r)rorse->rose(删除r)rose->ros(删除e)解题思路:动态规划(DP)✅状态定义dp[i]
- Visual Autoregressive Modeling: Scalable Image Generation via Next-Scale Prediction
zzfive
生成模型论文阅读kotlin开发语言android
论文链接:VisualAutoregressiveModeling:ScalableImageGenerationviaNext-ScalePrediction文章目录简介预测下一个token自回归模型范式分析VAR详解分词实现细节幂律缩放定律零样本泛化能力结论简介本文提出的视觉自回归建模/VAR这种新范式,其将图像的自回归学习重新定义为从粗到细的“下一个尺度预测”或“下一个分辨率预测”,与常规的
- 如何通过 WebSocket 接口订阅实时外汇行情数据(PHP 示例)
quant_1986
websocketphp网络协议开发语言后端网络金融
步骤1:准备工作确保已安装PHP和Composer安装WebSocket客户端库:composerrequiretextalk/websocket步骤2:编写代码订阅行情以下是最简可运行的PHP示例,订阅EUR/USD的1分钟K线数据:60]);//构造订阅请求$initMessage=["code"=>10004,"trace"=>uniqid(),"data"=>["arr"=>[["type
- 力扣 hot100 Day49
qq_51397044
Hot100leetcode算法数据结构
105.从前序与中序遍历序列构造二叉树给定两个整数数组preorder和inorder,其中preorder是二叉树的先序遍历,inorder是同一棵树的中序遍历,请构造二叉树并返回其根节点。//抄的classSolution{private:unordered_mapindex;TreeNode*myBuildTree(constvector&preorder,constvector&inord
- 力扣 hot100 Day44
qq_51397044
Hot100leetcode算法
98.验证二叉搜索树给你一个二叉树的根节点root,判断其是否是一个有效的二叉搜索树。有效二叉搜索树定义如下:节点的左子树只包含小于当前节点的数。节点的右子树只包含大于当前节点的数。所有左子树和右子树自身必须也是二叉搜索树//自己写的classSolution{public:voidinorderHelper(TreeNode*root,vector&result){if(root==nullpt
- 力扣 hot100 Day45
qq_51397044
Hot100leetcode算法
230.二叉搜索树中第K小的元素给定一个二叉搜索树的根节点root,和一个整数k,请你设计一个算法查找其中第k小的元素(从1开始计数)。//抄的classSolution{public:voidhelper(TreeNode*root,intk,int&count,int&result){if(!root)return;helper(root->left,k,count,result);count
- 在ComfyUI中CLIP Text Encode (Prompt)和CLIPTextEncodeFlux的区别
虎冯河
AIGCComfyUI
CLIPTextEncode(Prompt)CLIPTextEncodeFlux在ComfyUI中对token支持长度是否相同的详细技术对比:1、CLIPTextEncode(Prompt)通常来自:ComfyUI官方自带CLIPTextEncode节点。特点:✅使用OpenAICLIP模型(ViT-L/14等)✅默认最大支持77tokens(固定超参数)✅超过77tokens时:部分实现直接截断
- 我在黑马程序员学web前端
新手来了@click
前端
1网页由三部分组成1.、html负责网页的结构2.css、负责网页的美化,控制网页元素的样式3、js,负责网页交互html常见的标签:1、form表单input输入框select下拉菜单option下拉列表2、table表格thead表头tbody是表体tr行th表头加粗td是列br是换行2/CSS常见的三种引入方式行内样式、内部样式、外部样式用link关键字常用的元素选择器:标签选择器、id选择
- Leetcode703. 数据流中的第K大元素
LonnieQ
题目设计一个找到数据流中第K大元素的类(class)。注意是排序后的第K大元素,不是第K个不同的元素。你的KthLargest类需要一个同时接收整数k和整数数组nums的构造器,它包含数据流中的初始元素。每次调用KthLargest.add,返回当前数据流中第K大的元素。示例:intk=3;int[]arr=[4,5,8,2];KthLargestkthLargest=newKthLargest(
- Java-数构链表
2301_81674311
java链表开发语言
1.链表1.1链表的概念和结构链表是一种物理存储结构上非连续存储结构,数据元素的逻辑顺序是通过链表中引用链接次序实现的。这里大多讨论无头单向非循环链表。这种结构,结构简单,一般与其他数据结构结合,作为其他数据结构的子数据。1.2链表的实现publicclassMysingleList{staticclassListNode{publicintval;//节点的值域publicListNodenex
- 力扣 hot100 Day50
qq_51397044
Hot100leetcode算法职场和发展
437.路径总和III给定一个二叉树的根节点root,和一个整数targetSum,求该二叉树里节点值之和等于targetSum的路径的数目。路径不需要从根节点开始,也不需要在叶子节点结束,但是路径方向必须是向下的(只能从父节点到子节点)。//抄的classSolution{public:intpathSum(TreeNode*root,inttargetSum){unordered_mappre
- 浅谈EXT2文件系统----inode table
巭犇
文件系统linux数据库运维
Inodetable概述在EXT2文件系统中,inode表(InodeTable)是一个非常重要的结构,用于存储文件和目录的元数据。每个文件和目录都由一个inode(索引节点)来表示,inode中包含了关于该文件或目录的关键信息,如文件的大小、权限、所属用户、时间戳以及指向数据块的指针等。EXT2文件系统将所有inode结构集中存储在inode表中。内核源码structext2_inode{__l
- mysql主从数据同步
林鹤霄
mysql主从数据同步
配置mysql5.5主从服务器(转)
教程开始:一、安装MySQL
说明:在两台MySQL服务器192.168.21.169和192.168.21.168上分别进行如下操作,安装MySQL 5.5.22
二、配置MySQL主服务器(192.168.21.169)mysql -uroot -p &nb
- oracle学习笔记
caoyong
oracle
1、ORACLE的安装
a>、ORACLE的版本
8i,9i : i是internet
10g,11g : grid (网格)
12c : cloud (云计算)
b>、10g不支持win7
&
- 数据库,SQL零基础入门
天子之骄
sql数据库入门基本术语
数据库,SQL零基础入门
做网站肯定离不开数据库,本人之前没怎么具体接触SQL,这几天起早贪黑得各种入门,恶补脑洞。一些具体的知识点,可以让小白不再迷茫的术语,拿来与大家分享。
数据库,永久数据的一个或多个大型结构化集合,通常与更新和查询数据的软件相关
- pom.xml
一炮送你回车库
pom.xml
1、一级元素dependencies是可以被子项目继承的
2、一级元素dependencyManagement是定义该项目群里jar包版本号的,通常和一级元素properties一起使用,既然有继承,也肯定有一级元素modules来定义子元素
3、父项目里的一级元素<modules>
<module>lcas-admin-war</module>
<
- sql查地区省市县
3213213333332132
sqlmysql
-- db_yhm_city
SELECT * FROM db_yhm_city WHERE class_parent_id = 1 -- 海南 class_id = 9 港、奥、台 class_id = 33、34、35
SELECT * FROM db_yhm_city WHERE class_parent_id =169
SELECT d1.cla
- 关于监听器那些让人头疼的事
宝剑锋梅花香
画图板监听器鼠标监听器
本人初学JAVA,对于界面开发我只能说有点蛋疼,用JAVA来做界面的话确实需要一定的耐心(不使用插件,就算使用插件的话也没好多少)既然Java提供了界面开发,老师又要求做,只能硬着头皮上啦。但是监听器还真是个难懂的地方,我是上了几次课才略微搞懂了些。
- JAVA的遍历MAP
darkranger
map
Java Map遍历方式的选择
1. 阐述
对于Java中Map的遍历方式,很多文章都推荐使用entrySet,认为其比keySet的效率高很多。理由是:entrySet方法一次拿到所有key和value的集合;而keySet拿到的只是key的集合,针对每个key,都要去Map中额外查找一次value,从而降低了总体效率。那么实际情况如何呢?
为了解遍历性能的真实差距,包括在遍历ke
- POJ 2312 Battle City 优先多列+bfs
aijuans
搜索
来源:http://poj.org/problem?id=2312
题意:题目背景就是小时候玩的坦克大战,求从起点到终点最少需要多少步。已知S和R是不能走得,E是空的,可以走,B是砖,只有打掉后才可以通过。
思路:很容易看出来这是一道广搜的题目,但是因为走E和走B所需要的时间不一样,因此不能用普通的队列存点。因为对于走B来说,要先打掉砖才能通过,所以我们可以理解为走B需要两步,而走E是指需要1
- Hibernate与Jpa的关系,终于弄懂
avords
javaHibernate数据库jpa
我知道Jpa是一种规范,而Hibernate是它的一种实现。除了Hibernate,还有EclipseLink(曾经的toplink),OpenJPA等可供选择,所以使用Jpa的一个好处是,可以更换实现而不必改动太多代码。
在play中定义Model时,使用的是jpa的annotations,比如javax.persistence.Entity, Table, Column, OneToMany
- 酸爽的console.log
bee1314
console
在前端的开发中,console.log那是开发必备啊,简直直观。通过写小函数,组合大功能。更容易测试。但是在打版本时,就要删除console.log,打完版本进入开发状态又要添加,真不够爽。重复劳动太多。所以可以做些简单地封装,方便开发和上线。
/**
* log.js hufeng
* The safe wrapper for `console.xxx` functions
*
- 哈佛教授:穷人和过于忙碌的人有一个共同思维特质
bijian1013
时间管理励志人生穷人过于忙碌
一个跨学科团队今年完成了一项对资源稀缺状况下人的思维方式的研究,结论是:穷人和过于忙碌的人有一个共同思维特质,即注意力被稀缺资源过分占据,引起认知和判断力的全面下降。这项研究是心理学、行为经济学和政策研究学者协作的典范。
这个研究源于穆来纳森对自己拖延症的憎恨。他7岁从印度移民美国,很快就如鱼得水,哈佛毕业
- other operate
征客丶
OSosx
一、Mac Finder 设置排序方式,预览栏 在显示-》查看显示选项中
二、有时预览显示时,卡死在那,有可能是一些临时文件夹被删除了,如:/private/tmp[有待验证]
--------------------------------------------------------------------
若有其他凝问或文中有错误,请及时向我指出,
我好及时改正,同时也让我们一
- 【Scala五】分析Spark源代码总结的Scala语法三
bit1129
scala
1. If语句作为表达式
val properties = if (jobIdToActiveJob.contains(jobId)) {
jobIdToActiveJob(stage.jobId).properties
} else {
// this stage will be assigned to "default" po
- ZooKeeper 入门
BlueSkator
中间件zk
ZooKeeper是一个高可用的分布式数据管理与系统协调框架。基于对Paxos算法的实现,使该框架保证了分布式环境中数据的强一致性,也正是基于这样的特性,使得ZooKeeper解决很多分布式问题。网上对ZK的应用场景也有不少介绍,本文将结合作者身边的项目例子,系统地对ZK的应用场景进行一个分门归类的介绍。
值得注意的是,ZK并非天生就是为这些应用场景设计的,都是后来众多开发者根据其框架的特性,利
- MySQL取得当前时间的函数是什么 格式化日期的函数是什么
BreakingBad
mysqlDate
取得当前时间用 now() 就行。
在数据库中格式化时间 用DATE_FORMA T(date, format) .
根据格式串format 格式化日期或日期和时间值date,返回结果串。
可用DATE_FORMAT( ) 来格式化DATE 或DATETIME 值,以便得到所希望的格式。根据format字符串格式化date值:
%S, %s 两位数字形式的秒( 00,01,
- 读《研磨设计模式》-代码笔记-组合模式
bylijinnan
java设计模式
声明: 本文只为方便我个人查阅和理解,详细的分析以及源代码请移步 原作者的博客http://chjavach.iteye.com/
import java.util.ArrayList;
import java.util.List;
abstract class Component {
public abstract void printStruct(Str
- 4_JAVA+Oracle面试题(有答案)
chenke
oracle
基础测试题
卷面上不能出现任何的涂写文字,所有的答案要求写在答题纸上,考卷不得带走。
选择题
1、 What will happen when you attempt to compile and run the following code? (3)
public class Static {
static {
int x = 5; // 在static内有效
}
st
- 新一代工作流系统设计目标
comsci
工作算法脚本
用户只需要给工作流系统制定若干个需求,流程系统根据需求,并结合事先输入的组织机构和权限结构,调用若干算法,在流程展示版面上面显示出系统自动生成的流程图,然后由用户根据实际情况对该流程图进行微调,直到满意为止,流程在运行过程中,系统和用户可以根据情况对流程进行实时的调整,包括拓扑结构的调整,权限的调整,内置脚本的调整。。。。。
在这个设计中,最难的地方是系统根据什么来生成流
- oracle 行链接与行迁移
daizj
oracle行迁移
表里的一行对于一个数据块太大的情况有二种(一行在一个数据块里放不下)
第一种情况:
INSERT的时候,INSERT时候行的大小就超一个块的大小。Oracle把这行的数据存储在一连串的数据块里(Oracle Stores the data for the row in a chain of data blocks),这种情况称为行链接(Row Chain),一般不可避免(除非使用更大的数据
- [JShop]开源电子商务系统jshop的系统缓存实现
dinguangx
jshop电子商务
前言
jeeshop中通过SystemManager管理了大量的缓存数据,来提升系统的性能,但这些缓存数据全部都是存放于内存中的,无法满足特定场景的数据更新(如集群环境)。JShop对jeeshop的缓存机制进行了扩展,提供CacheProvider来辅助SystemManager管理这些缓存数据,通过CacheProvider,可以把缓存存放在内存,ehcache,redis,memcache
- 初三全学年难记忆单词
dcj3sjt126com
englishword
several 儿子;若干
shelf 架子
knowledge 知识;学问
librarian 图书管理员
abroad 到国外,在国外
surf 冲浪
wave 浪;波浪
twice 两次;两倍
describe 描写;叙述
especially 特别;尤其
attract 吸引
prize 奖品;奖赏
competition 比赛;竞争
event 大事;事件
O
- sphinx实践
dcj3sjt126com
sphinx
安装参考地址:http://briansnelson.com/How_to_install_Sphinx_on_Centos_Server
yum install sphinx
如果失败的话使用下面的方式安装
wget http://sphinxsearch.com/files/sphinx-2.2.9-1.rhel6.x86_64.rpm
yum loca
- JPA之JPQL(三)
frank1234
ormjpaJPQL
1 什么是JPQL
JPQL是Java Persistence Query Language的简称,可以看成是JPA中的HQL, JPQL支持各种复杂查询。
2 检索单个对象
@Test
public void querySingleObject1() {
Query query = em.createQuery("sele
- Remove Duplicates from Sorted Array II
hcx2013
remove
Follow up for "Remove Duplicates":What if duplicates are allowed at most twice?
For example,Given sorted array nums = [1,1,1,2,2,3],
Your function should return length
- Spring4新特性——Groovy Bean定义DSL
jinnianshilongnian
spring 4
Spring4新特性——泛型限定式依赖注入
Spring4新特性——核心容器的其他改进
Spring4新特性——Web开发的增强
Spring4新特性——集成Bean Validation 1.1(JSR-349)到SpringMVC
Spring4新特性——Groovy Bean定义DSL
Spring4新特性——更好的Java泛型操作API
Spring4新
- CentOS安装Mysql5.5
liuxingguome
centos
CentOS下以RPM方式安装MySQL5.5
首先卸载系统自带Mysql:
yum remove mysql mysql-server mysql-libs compat-mysql51
rm -rf /var/lib/mysql
rm /etc/my.cnf
查看是否还有mysql软件:
rpm -qa|grep mysql
去http://dev.mysql.c
- 第14章 工具函数(下)
onestopweb
函数
index.html
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/
- POJ 1050
SaraWon
二维数组子矩阵最大和
POJ ACM第1050题的详细描述,请参照
http://acm.pku.edu.cn/JudgeOnline/problem?id=1050
题目意思:
给定包含有正负整型的二维数组,找出所有子矩阵的和的最大值。
如二维数组
0 -2 -7 0
9 2 -6 2
-4 1 -4 1
-1 8 0 -2
中和最大的子矩阵是
9 2
-4 1
-1 8
且最大和是15
- [5]设计模式——单例模式
tsface
java单例设计模式虚拟机
单例模式:保证一个类仅有一个实例,并提供一个访问它的全局访问点
安全的单例模式:
/*
* @(#)Singleton.java 2014-8-1
*
* Copyright 2014 XXXX, Inc. All rights reserved.
*/
package com.fiberhome.singleton;
- Java8全新打造,英语学习supertool
yangshangchuan
javasuperword闭包java8函数式编程
superword是一个Java实现的英文单词分析软件,主要研究英语单词音近形似转化规律、前缀后缀规律、词之间的相似性规律等等。Clean code、Fluent style、Java8 feature: Lambdas, Streams and Functional-style Programming。
升学考试、工作求职、充电提高,都少不了英语的身影,英语对我们来说实在太重要