无监督参考图像评价-自然图像评价指标-NIQE python代码 可运行
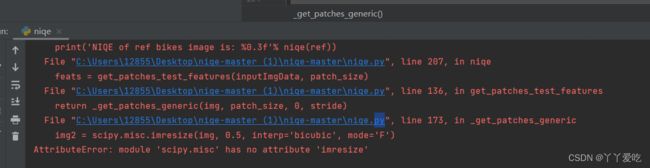
github上下载的源码NIQE出现imresize、imread导入错误问题。
解决方法
将博主最后提供的代码复制粘贴到niqe代码里面成功解决。
贴出代码:
需要python3.6版本
import numpy as np
import scipy.misc
import scipy.io
from os.path import dirname
from os.path import join
import scipy
from PIL import Image
import numpy as np
import scipy.ndimage
import numpy as np
import scipy.special
import math
#需要python3.6版本
'''
Code for downloading and processing Caltech Pedestrian Dataset - test part (P. Dollar et al. 2009, http://www.vision.caltech.edu/Image_Datasets/CaltechPedestrians/).
Based on code related to PredNet - Lotter et al. 2016 (https://arxiv.org/abs/1605.08104 https://github.com/coxlab/prednet).
Method of resizing was specified (bicubic).
'''
"""
A collection of image utilities using the Python Imaging Library (PIL).
Note that PIL is not a dependency of SciPy and this module is not
available on systems that don't have PIL installed.
"""
# from __future__ import division, print_function, absolute_import
# Functions which need the PIL
import numpy
import tempfile
from numpy import (amin, amax, ravel, asarray, arange, ones, newaxis,
transpose, iscomplexobj, uint8, issubdtype, array)
try:
from PIL import Image, ImageFilter
except ImportError:
import Image
import ImageFilter
if not hasattr(Image, 'frombytes'):
Image.frombytes = Image.fromstring
__all__ = ['fromimage', 'toimage', 'imsave', 'imread', 'bytescale',
'imrotate', 'imresize', 'imshow', 'imfilter']
@numpy.deprecate(message="`bytescale` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.")
def bytescale(data, cmin=None, cmax=None, high=255, low=0):
"""
Byte scales an array (image).
Byte scaling means converting the input image to uint8 dtype and scaling
the range to ``(low, high)`` (default 0-255).
If the input image already has dtype uint8, no scaling is done.
This function is only available if Python Imaging Library (PIL) is installed.
Parameters
----------
data : ndarray
PIL image data array.
cmin : scalar, optional
Bias scaling of small values. Default is ``data.min()``.
cmax : scalar, optional
Bias scaling of large values. Default is ``data.max()``.
high : scalar, optional
Scale max value to `high`. Default is 255.
low : scalar, optional
Scale min value to `low`. Default is 0.
Returns
-------
img_array : uint8 ndarray
The byte-scaled array.
Examples
--------
>>> from scipy.misc import bytescale
>>> img = np.array([[ 91.06794177, 3.39058326, 84.4221549 ],
... [ 73.88003259, 80.91433048, 4.88878881],
... [ 51.53875334, 34.45808177, 27.5873488 ]])
>>> bytescale(img)
array([[255, 0, 236],
[205, 225, 4],
[140, 90, 70]], dtype=uint8)
>>> bytescale(img, high=200, low=100)
array([[200, 100, 192],
[180, 188, 102],
[155, 135, 128]], dtype=uint8)
>>> bytescale(img, cmin=0, cmax=255)
array([[91, 3, 84],
[74, 81, 5],
[52, 34, 28]], dtype=uint8)
"""
if data.dtype == uint8:
return data
if high > 255:
raise ValueError("`high` should be less than or equal to 255.")
if low < 0:
raise ValueError("`low` should be greater than or equal to 0.")
if high < low:
raise ValueError("`high` should be greater than or equal to `low`.")
if cmin is None:
cmin = data.min()
if cmax is None:
cmax = data.max()
cscale = cmax - cmin
if cscale < 0:
raise ValueError("`cmax` should be larger than `cmin`.")
elif cscale == 0:
cscale = 1
scale = float(high - low) / cscale
bytedata = (data - cmin) * scale + low
return (bytedata.clip(low, high) + 0.5).astype(uint8)
@numpy.deprecate(message="`imread` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use ``imageio.imread`` instead.")
def imread(name, flatten=False, mode=None):
"""
Read an image from a file as an array.
This function is only available if Python Imaging Library (PIL) is installed.
Parameters
----------
name : str or file object
The file name or file object to be read.
flatten : bool, optional
If True, flattens the color layers into a single gray-scale layer.
mode : str, optional
Mode to convert image to, e.g. ``'RGB'``. See the Notes for more
details.
Returns
-------
imread : ndarray
The array obtained by reading the image.
Notes
-----
`imread` uses the Python Imaging Library (PIL) to read an image.
The following notes are from the PIL documentation.
`mode` can be one of the following strings:
* 'L' (8-bit pixels, black and white)
* 'P' (8-bit pixels, mapped to any other mode using a color palette)
* 'RGB' (3x8-bit pixels, true color)
* 'RGBA' (4x8-bit pixels, true color with transparency mask)
* 'CMYK' (4x8-bit pixels, color separation)
* 'YCbCr' (3x8-bit pixels, color video format)
* 'I' (32-bit signed integer pixels)
* 'F' (32-bit floating point pixels)
PIL also provides limited support for a few special modes, including
'LA' ('L' with alpha), 'RGBX' (true color with padding) and 'RGBa'
(true color with premultiplied alpha).
When translating a color image to black and white (mode 'L', 'I' or
'F'), the library uses the ITU-R 601-2 luma transform::
L = R * 299/1000 + G * 587/1000 + B * 114/1000
When `flatten` is True, the image is converted using mode 'F'.
When `mode` is not None and `flatten` is True, the image is first
converted according to `mode`, and the result is then flattened using
mode 'F'.
"""
im = Image.open(name)
return fromimage(im, flatten=flatten, mode=mode)
@numpy.deprecate(message="`imsave` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use ``imageio.imwrite`` instead.")
def imsave(name, arr, format=None):
"""
Save an array as an image.
This function is only available if Python Imaging Library (PIL) is installed.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Parameters
----------
name : str or file object
Output file name or file object.
arr : ndarray, MxN or MxNx3 or MxNx4
Array containing image values. If the shape is ``MxN``, the array
represents a grey-level image. Shape ``MxNx3`` stores the red, green
and blue bands along the last dimension. An alpha layer may be
included, specified as the last colour band of an ``MxNx4`` array.
format : str
Image format. If omitted, the format to use is determined from the
file name extension. If a file object was used instead of a file name,
this parameter should always be used.
Examples
--------
Construct an array of gradient intensity values and save to file:
>>> from scipy.misc import imsave
>>> x = np.zeros((255, 255))
>>> x = np.zeros((255, 255), dtype=np.uint8)
>>> x[:] = np.arange(255)
>>> imsave('gradient.png', x)
Construct an array with three colour bands (R, G, B) and store to file:
>>> rgb = np.zeros((255, 255, 3), dtype=np.uint8)
>>> rgb[..., 0] = np.arange(255)
>>> rgb[..., 1] = 55
>>> rgb[..., 2] = 1 - np.arange(255)
>>> imsave('rgb_gradient.png', rgb)
"""
im = toimage(arr, channel_axis=2)
if format is None:
im.save(name)
else:
im.save(name, format)
return
@numpy.deprecate(message="`fromimage` is deprecated in SciPy 1.0.0. "
"and will be removed in 1.2.0.\n"
"Use ``np.asarray(im)`` instead.")
def fromimage(im, flatten=False, mode=None):
"""
Return a copy of a PIL image as a numpy array.
This function is only available if Python Imaging Library (PIL) is installed.
Parameters
----------
im : PIL image
Input image.
flatten : bool
If true, convert the output to grey-scale.
mode : str, optional
Mode to convert image to, e.g. ``'RGB'``. See the Notes of the
`imread` docstring for more details.
Returns
-------
fromimage : ndarray
The different colour bands/channels are stored in the
third dimension, such that a grey-image is MxN, an
RGB-image MxNx3 and an RGBA-image MxNx4.
"""
if not Image.isImageType(im):
raise TypeError("Input is not a PIL image.")
if mode is not None:
if mode != im.mode:
im = im.convert(mode)
elif im.mode == 'P':
# Mode 'P' means there is an indexed "palette". If we leave the mode
# as 'P', then when we do `a = array(im)` below, `a` will be a 2-D
# containing the indices into the palette, and not a 3-D array
# containing the RGB or RGBA values.
if 'transparency' in im.info:
im = im.convert('RGBA')
else:
im = im.convert('RGB')
if flatten:
im = im.convert('F')
elif im.mode == '1':
# Workaround for crash in PIL. When im is 1-bit, the call array(im)
# can cause a seg. fault, or generate garbage. See
# https://github.com/scipy/scipy/issues/2138 and
# https://github.com/python-pillow/Pillow/issues/350.
#
# This converts im from a 1-bit image to an 8-bit image.
im = im.convert('L')
a = array(im)
return a
_errstr = "Mode is unknown or incompatible with input array shape."
@numpy.deprecate(message="`toimage` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use Pillow's ``Image.fromarray`` directly instead.")
def toimage(arr, high=255, low=0, cmin=None, cmax=None, pal=None,
mode=None, channel_axis=None):
"""Takes a numpy array and returns a PIL image.
This function is only available if Python Imaging Library (PIL) is installed.
The mode of the PIL image depends on the array shape and the `pal` and
`mode` keywords.
For 2-D arrays, if `pal` is a valid (N,3) byte-array giving the RGB values
(from 0 to 255) then ``mode='P'``, otherwise ``mode='L'``, unless mode
is given as 'F' or 'I' in which case a float and/or integer array is made.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Notes
-----
For 3-D arrays, the `channel_axis` argument tells which dimension of the
array holds the channel data.
For 3-D arrays if one of the dimensions is 3, the mode is 'RGB'
by default or 'YCbCr' if selected.
The numpy array must be either 2 dimensional or 3 dimensional.
"""
data = asarray(arr)
if iscomplexobj(data):
raise ValueError("Cannot convert a complex-valued array.")
shape = list(data.shape)
valid = len(shape) == 2 or ((len(shape) == 3) and
((3 in shape) or (4 in shape)))
if not valid:
raise ValueError("'arr' does not have a suitable array shape for "
"any mode.")
if len(shape) == 2:
shape = (shape[1], shape[0]) # columns show up first
if mode == 'F':
data32 = data.astype(numpy.float32)
image = Image.frombytes(mode, shape, data32.tostring())
return image
if mode in [None, 'L', 'P']:
bytedata = bytescale(data, high=high, low=low,
cmin=cmin, cmax=cmax)
image = Image.frombytes('L', shape, bytedata.tostring())
if pal is not None:
image.putpalette(asarray(pal, dtype=uint8).tostring())
# Becomes a mode='P' automagically.
elif mode == 'P': # default gray-scale
pal = (arange(0, 256, 1, dtype=uint8)[:, newaxis] *
ones((3,), dtype=uint8)[newaxis, :])
image.putpalette(asarray(pal, dtype=uint8).tostring())
return image
if mode == '1': # high input gives threshold for 1
bytedata = (data > high)
image = Image.frombytes('1', shape, bytedata.tostring())
return image
if cmin is None:
cmin = amin(ravel(data))
if cmax is None:
cmax = amax(ravel(data))
data = (data * 1.0 - cmin) * (high - low) / (cmax - cmin) + low
if mode == 'I':
data32 = data.astype(numpy.uint32)
image = Image.frombytes(mode, shape, data32.tostring())
else:
raise ValueError(_errstr)
return image
# if here then 3-d array with a 3 or a 4 in the shape length.
# Check for 3 in datacube shape --- 'RGB' or 'YCbCr'
if channel_axis is None:
if (3 in shape):
ca = numpy.flatnonzero(asarray(shape) == 3)[0]
else:
ca = numpy.flatnonzero(asarray(shape) == 4)
if len(ca):
ca = ca[0]
else:
raise ValueError("Could not find channel dimension.")
else:
ca = channel_axis
numch = shape[ca]
if numch not in [3, 4]:
raise ValueError("Channel axis dimension is not valid.")
bytedata = bytescale(data, high=high, low=low, cmin=cmin, cmax=cmax)
if ca == 2:
strdata = bytedata.tostring()
shape = (shape[1], shape[0])
elif ca == 1:
strdata = transpose(bytedata, (0, 2, 1)).tostring()
shape = (shape[2], shape[0])
elif ca == 0:
strdata = transpose(bytedata, (1, 2, 0)).tostring()
shape = (shape[2], shape[1])
if mode is None:
if numch == 3:
mode = 'RGB'
else:
mode = 'RGBA'
if mode not in ['RGB', 'RGBA', 'YCbCr', 'CMYK']:
raise ValueError(_errstr)
if mode in ['RGB', 'YCbCr']:
if numch != 3:
raise ValueError("Invalid array shape for mode.")
if mode in ['RGBA', 'CMYK']:
if numch != 4:
raise ValueError("Invalid array shape for mode.")
# Here we know data and mode is correct
image = Image.frombytes(mode, shape, strdata)
return image
@numpy.deprecate(message="`imrotate` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use ``skimage.transform.rotate`` instead.")
def imrotate(arr, angle, interp='bilinear'):
"""
Rotate an image counter-clockwise by angle degrees.
This function is only available if Python Imaging Library (PIL) is installed.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Parameters
----------
arr : ndarray
Input array of image to be rotated.
angle : float
The angle of rotation.
interp : str, optional
Interpolation
- 'nearest' : for nearest neighbor
- 'bilinear' : for bilinear
- 'lanczos' : for lanczos
- 'cubic' : for bicubic
- 'bicubic' : for bicubic
Returns
-------
imrotate : ndarray
The rotated array of image.
"""
arr = asarray(arr)
func = {'nearest': 0, 'lanczos': 1, 'bilinear': 2, 'bicubic': 3, 'cubic': 3}
im = toimage(arr)
im = im.rotate(angle, resample=func[interp])
return fromimage(im)
@numpy.deprecate(message="`imshow` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use ``matplotlib.pyplot.imshow`` instead.")
def imshow(arr):
"""
Simple showing of an image through an external viewer.
This function is only available if Python Imaging Library (PIL) is installed.
Uses the image viewer specified by the environment variable
SCIPY_PIL_IMAGE_VIEWER, or if that is not defined then `see`,
to view a temporary file generated from array data.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Parameters
----------
arr : ndarray
Array of image data to show.
Returns
-------
None
Examples
--------
>>> a = np.tile(np.arange(255), (255,1))
>>> from scipy import misc
>>> misc.imshow(a)
"""
im = toimage(arr)
fnum, fname = tempfile.mkstemp('.png')
try:
im.save(fname)
except Exception:
raise RuntimeError("Error saving temporary image data.")
import os
os.close(fnum)
cmd = os.environ.get('SCIPY_PIL_IMAGE_VIEWER', 'see')
status = os.system("%s %s" % (cmd, fname))
os.unlink(fname)
if status != 0:
raise RuntimeError('Could not execute image viewer.')
@numpy.deprecate(message="`imresize` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.3.0.\n"
"Use Pillow instead: ``numpy.array(Image.fromarray(arr).resize())``.")
def imresize(arr, size, interp='bilinear', mode=None):
"""
Resize an image.
This function is only available if Python Imaging Library (PIL) is installed.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Parameters
----------
arr : ndarray
The array of image to be resized.
size : int, float or tuple
* int - Percentage of current size.
* float - Fraction of current size.
* tuple - Size of the output image (height, width).
interp : str, optional
Interpolation to use for re-sizing ('nearest', 'lanczos', 'bilinear',
'bicubic' or 'cubic').
mode : str, optional
The PIL image mode ('P', 'L', etc.) to convert `arr` before resizing.
If ``mode=None`` (the default), 2-D images will be treated like
``mode='L'``, i.e. casting to long integer. For 3-D and 4-D arrays,
`mode` will be set to ``'RGB'`` and ``'RGBA'`` respectively.
Returns
-------
imresize : ndarray
The resized array of image.
See Also
--------
toimage : Implicitly used to convert `arr` according to `mode`.
scipy.ndimage.zoom : More generic implementation that does not use PIL.
"""
im = toimage(arr, mode=mode)
ts = type(size)
if issubdtype(ts, numpy.signedinteger):
percent = size / 100.0
size = tuple((array(im.size) * percent).astype(int))
elif issubdtype(type(size), numpy.floating):
size = tuple((array(im.size) * size).astype(int))
else:
size = (size[1], size[0])
func = {'nearest': 0, 'lanczos': 1, 'bilinear': 2, 'bicubic': 3, 'cubic': 3}
imnew = im.resize(size, resample=func[interp])
return fromimage(imnew)
@numpy.deprecate(message="`imfilter` is deprecated in SciPy 1.0.0, "
"and will be removed in 1.2.0.\n"
"Use Pillow filtering functionality directly.")
def imfilter(arr, ftype):
"""
Simple filtering of an image.
This function is only available if Python Imaging Library (PIL) is installed.
.. warning::
This function uses `bytescale` under the hood to rescale images to use
the full (0, 255) range if ``mode`` is one of ``None, 'L', 'P', 'l'``.
It will also cast data for 2-D images to ``uint32`` for ``mode=None``
(which is the default).
Parameters
----------
arr : ndarray
The array of Image in which the filter is to be applied.
ftype : str
The filter that has to be applied. Legal values are:
'blur', 'contour', 'detail', 'edge_enhance', 'edge_enhance_more',
'emboss', 'find_edges', 'smooth', 'smooth_more', 'sharpen'.
Returns
-------
imfilter : ndarray
The array with filter applied.
Raises
------
ValueError
*Unknown filter type.* If the filter you are trying
to apply is unsupported.
"""
_tdict = {'blur': ImageFilter.BLUR,
'contour': ImageFilter.CONTOUR,
'detail': ImageFilter.DETAIL,
'edge_enhance': ImageFilter.EDGE_ENHANCE,
'edge_enhance_more': ImageFilter.EDGE_ENHANCE_MORE,
'emboss': ImageFilter.EMBOSS,
'find_edges': ImageFilter.FIND_EDGES,
'smooth': ImageFilter.SMOOTH,
'smooth_more': ImageFilter.SMOOTH_MORE,
'sharpen': ImageFilter.SHARPEN
}
im = toimage(arr)
if ftype not in _tdict:
raise ValueError("Unknown filter type.")
return fromimage(im.filter(_tdict[ftype]))
# 下面加上自己需要的文件,也可以放在另一个文件,导入该函数
gamma_range = np.arange(0.2, 10, 0.001)
a = scipy.special.gamma(2.0/gamma_range)
a *= a
b = scipy.special.gamma(1.0/gamma_range)
c = scipy.special.gamma(3.0/gamma_range)
prec_gammas = a/(b*c)
def aggd_features(imdata):
#flatten imdata
imdata.shape = (len(imdata.flat),)
imdata2 = imdata*imdata
left_data = imdata2[imdata<0]
right_data = imdata2[imdata>=0]
left_mean_sqrt = 0
right_mean_sqrt = 0
if len(left_data) > 0:
left_mean_sqrt = np.sqrt(np.average(left_data))
if len(right_data) > 0:
right_mean_sqrt = np.sqrt(np.average(right_data))
if right_mean_sqrt != 0:
gamma_hat = left_mean_sqrt/right_mean_sqrt
else:
gamma_hat = np.inf
#solve r-hat norm
imdata2_mean = np.mean(imdata2)
if imdata2_mean != 0:
r_hat = (np.average(np.abs(imdata))**2) / (np.average(imdata2))
else:
r_hat = np.inf
rhat_norm = r_hat * (((math.pow(gamma_hat, 3) + 1)*(gamma_hat + 1)) / math.pow(math.pow(gamma_hat, 2) + 1, 2))
#solve alpha by guessing values that minimize ro
pos = np.argmin((prec_gammas - rhat_norm)**2);
alpha = gamma_range[pos]
gam1 = scipy.special.gamma(1.0/alpha)
gam2 = scipy.special.gamma(2.0/alpha)
gam3 = scipy.special.gamma(3.0/alpha)
aggdratio = np.sqrt(gam1) / np.sqrt(gam3)
bl = aggdratio * left_mean_sqrt
br = aggdratio * right_mean_sqrt
#mean parameter
N = (br - bl)*(gam2 / gam1)#*aggdratio
return (alpha, N, bl, br, left_mean_sqrt, right_mean_sqrt)
def ggd_features(imdata):
nr_gam = 1/prec_gammas
sigma_sq = np.var(imdata)
E = np.mean(np.abs(imdata))
rho = sigma_sq/E**2
pos = np.argmin(np.abs(nr_gam - rho));
return gamma_range[pos], sigma_sq
def paired_product(new_im):
shift1 = np.roll(new_im.copy(), 1, axis=1)
shift2 = np.roll(new_im.copy(), 1, axis=0)
shift3 = np.roll(np.roll(new_im.copy(), 1, axis=0), 1, axis=1)
shift4 = np.roll(np.roll(new_im.copy(), 1, axis=0), -1, axis=1)
H_img = shift1 * new_im
V_img = shift2 * new_im
D1_img = shift3 * new_im
D2_img = shift4 * new_im
return (H_img, V_img, D1_img, D2_img)
def gen_gauss_window(lw, sigma):
sd = np.float32(sigma)
lw = int(lw)
weights = [0.0] * (2 * lw + 1)
weights[lw] = 1.0
sum = 1.0
sd *= sd
for ii in range(1, lw + 1):
tmp = np.exp(-0.5 * np.float32(ii * ii) / sd)
weights[lw + ii] = tmp
weights[lw - ii] = tmp
sum += 2.0 * tmp
for ii in range(2 * lw + 1):
weights[ii] /= sum
return weights
def compute_image_mscn_transform(image, C=1, avg_window=None, extend_mode='constant'):
if avg_window is None:
avg_window = gen_gauss_window(3, 7.0/6.0)
assert len(np.shape(image)) == 2
h, w = np.shape(image)
mu_image = np.zeros((h, w), dtype=np.float32)
var_image = np.zeros((h, w), dtype=np.float32)
image = np.array(image).astype('float32')
scipy.ndimage.correlate1d(image, avg_window, 0, mu_image, mode=extend_mode)
scipy.ndimage.correlate1d(mu_image, avg_window, 1, mu_image, mode=extend_mode)
scipy.ndimage.correlate1d(image**2, avg_window, 0, var_image, mode=extend_mode)
scipy.ndimage.correlate1d(var_image, avg_window, 1, var_image, mode=extend_mode)
var_image = np.sqrt(np.abs(var_image - mu_image**2))
return (image - mu_image)/(var_image + C), var_image, mu_image
def _niqe_extract_subband_feats(mscncoefs):
# alpha_m, = extract_ggd_features(mscncoefs)
alpha_m, N, bl, br, lsq, rsq = aggd_features(mscncoefs.copy())
pps1, pps2, pps3, pps4 = paired_product(mscncoefs)
alpha1, N1, bl1, br1, lsq1, rsq1 = aggd_features(pps1)
alpha2, N2, bl2, br2, lsq2, rsq2 = aggd_features(pps2)
alpha3, N3, bl3, br3, lsq3, rsq3 = aggd_features(pps3)
alpha4, N4, bl4, br4, lsq4, rsq4 = aggd_features(pps4)
return np.array([alpha_m, (bl+br)/2.0,
alpha1, N1, bl1, br1, # (V)
alpha2, N2, bl2, br2, # (H)
alpha3, N3, bl3, bl3, # (D1)
alpha4, N4, bl4, bl4, # (D2)
])
def get_patches_train_features(img, patch_size, stride=8):
return _get_patches_generic(img, patch_size, 1, stride)
def get_patches_test_features(img, patch_size, stride=8):
return _get_patches_generic(img, patch_size, 0, stride)
def extract_on_patches(img, patch_size):
h, w = img.shape
patch_size = np.int(patch_size)
patches = []
for j in range(0, h-patch_size+1, patch_size):
for i in range(0, w-patch_size+1, patch_size):
patch = img[j:j+patch_size, i:i+patch_size]
patches.append(patch)
patches = np.array(patches)
patch_features = []
for p in patches:
patch_features.append(_niqe_extract_subband_feats(p))
patch_features = np.array(patch_features)
return patch_features
def _get_patches_generic(img, patch_size, is_train, stride):
h, w = np.shape(img)
if h < patch_size or w < patch_size:
print("Input image is too small")
exit(0)
# ensure that the patch divides evenly into img
hoffset = (h % patch_size)
woffset = (w % patch_size)
if hoffset > 0:
img = img[:-hoffset, :]
if woffset > 0:
img = img[:, :-woffset]
img = img.astype(np.float32)
img2 = scipy.misc.imresize(img, 0.5, interp='bicubic', mode='F')
mscn1, var, mu = compute_image_mscn_transform(img)
mscn1 = mscn1.astype(np.float32)
mscn2, _, _ = compute_image_mscn_transform(img2)
mscn2 = mscn2.astype(np.float32)
feats_lvl1 = extract_on_patches(mscn1, patch_size)
feats_lvl2 = extract_on_patches(mscn2, patch_size/2)
feats = np.hstack((feats_lvl1, feats_lvl2))# feats_lvl3))
return feats
def niqe(inputImgData):
patch_size = 96
module_path = dirname(__file__)
# TODO: memoize
params = scipy.io.loadmat(join(module_path, 'data', 'niqe_image_params.mat'))
pop_mu = np.ravel(params["pop_mu"])
pop_cov = params["pop_cov"]
M, N = inputImgData.shape
# assert C == 1, "niqe called with videos containing %d channels. Please supply only the luminance channel" % (C,)
assert M > (patch_size*2+1), "niqe called with small frame size, requires > 192x192 resolution video using current training parameters"
assert N > (patch_size*2+1), "niqe called with small frame size, requires > 192x192 resolution video using current training parameters"
feats = get_patches_test_features(inputImgData, patch_size)
sample_mu = np.mean(feats, axis=0)
sample_cov = np.cov(feats.T)
X = sample_mu - pop_mu
covmat = ((pop_cov+sample_cov)/2.0)
pinvmat = scipy.linalg.pinv(covmat)
niqe_score = np.sqrt(np.dot(np.dot(X, pinvmat), X))
return niqe_score
if __name__ == "__main__":
ref = np.array(Image.open('./test_imgs/bikes.bmp').convert('LA'))[:,:,0] # ref
dis = np.array(Image.open('./test_imgs/bikes_distorted.bmp').convert('LA'))[:,:,0] # dis
print('NIQE of ref bikes image is: %0.3f'% niqe(ref))
print('NIQE of dis bikes image is: %0.3f'% niqe(dis))
ref = np.array(Image.open('./test_imgs/parrots.bmp').convert('LA'))[:,:,0] # ref
dis = np.array(Image.open('./test_imgs/parrots_distorted.bmp').convert('LA'))[:,:,0] # dis
print('NIQE of ref parrot image is: %0.3f'% niqe(ref))
print('NIQE of dis parrot image is: %0.3f'% niqe(dis))