【Matlab】基于遗传算法优化 BP 神经网络的时间序列预测(Excel可直接替换数据)
【Matlab】基于遗传算法优化 BP 神经网络的时间序列预测(Excel可直接替换数据)
- 1.模型原理
- 2.文件结构
- 3.Excel数据
- 4.分块代码
-
- 4.1 arithXover.m
- 4.2 delta.m
- 4.3 ga.m
- 4.4 gabpEval.m
- 4.5 initializega.m
- 4.6 maxGenTerm.m
- 4.7 nonUnifMutation.m
- 4.8 normGeomSelect.m
- 4.9 parse.m
- 4.10 gadecod.m
- 4.11 main.m
- 5.运行结果
1.模型原理
当遗传算法用于优化BP神经网络的时间序列预测时,我们可以使用如下的数学原理来描述其步骤:
-
定义问题:
假设我们有一个时间序列数据集 { ( x 1 , y 1 ) , ( x 2 , y 2 ) , … , ( x n , y n ) } \{(x_1, y_1), (x_2, y_2), \ldots, (x_n, y_n)\} {(x1,y1),(x2,y2),…,(xn,yn)},其中 x i x_i xi 是时间步i的输入样本, y i y_i yi 是对应的时间步i的目标值(实际输出)。时间序列预测任务的目标是根据过去的观测值预测未来的值。 -
BP神经网络结构:
假设BP神经网络有L层,第l层有 n l n_l nl 个神经元。 w i j ( l ) w_{ij}^{(l)} wij(l) 表示第l-1层第i个神经元到第l层第j个神经元之间的连接权重, b i ( l ) b_i^{(l)} bi(l) 表示第l层第i个神经元的偏置。神经网络的输入是 x i x_i xi,第l层第i个神经元的输出(经过激活函数后的值)为 a i ( l ) a_i^{(l)} ai(l)。 -
目标函数:
在时间序列预测问题中,我们可以使用均方误差(Mean Squared Error,MSE)作为BP神经网络的目标函数,用于衡量实际输出值与预测输出值之间的差距。MSE可以定义为:MSE = 1 n ∑ i = 1 n ( y i − y ^ i ) 2 \text{MSE} = \frac{1}{n} \sum_{i=1}^{n} (y_i - \hat{y}_i)^2 MSE=n1i=1∑n(yi−y^i)2
其中, n n n是时间序列的长度, y i y_i yi是第i个时间步的实际输出, y ^ i \hat{y}_i y^i是BP神经网络在输入 x i x_i xi 上的预测输出。
-
遗传算法编码:
同样地,我们将BP神经网络的所有参数(权重和偏置)编码成一个染色体,通常用一个长向量来表示。染色体可以表示为:染色体 = [ w 11 ( 1 ) , w 12 ( 1 ) , … , w n 1 ( 1 ) ( 1 ) , w 11 ( 2 ) , w 12 ( 2 ) , … , w n L − 1 ( L ) ( L ) , b 1 ( 1 ) , b 2 ( 1 ) , … , b n 1 ( L ) ( L ) ] \text{染色体} = [w_{11}^{(1)}, w_{12}^{(1)}, \ldots, w_{n_1^{(1)}}^{(1)}, w_{11}^{(2)}, w_{12}^{(2)}, \ldots, w_{n_{L-1}^{(L)}}^{(L)}, b_1^{(1)}, b_2^{(1)}, \ldots, b_{n_1^{(L)}}^{(L)}] 染色体=[w11(1),w12(1),…,wn1(1)(1),w11(2),w12(2),…,wnL−1(L)(L),b1(1),b2(1),…,bn1(L)(L)]
其中, w i j ( l ) w_{ij}^{(l)} wij(l) 表示第l-1层第i个神经元到第l层第j个神经元之间的连接权重, b i ( l ) b_i^{(l)} bi(l) 表示第l-1层第i个神经元的偏置。
-
适应度函数:
在时间序列预测问题中,适应度函数可以定义为目标函数MSE的倒数(加上一个常数,以避免除以0):适应度 = 1 MSE + ϵ \text{适应度} = \frac{1}{\text{MSE} + \epsilon} 适应度=MSE+ϵ1
其中, ϵ \epsilon ϵ 是一个小的常数,用于避免分母为0的情况。
-
选择:
采用基于适应度函数的选择策略,选择适应度较高的个体作为“父代”,用于产生下一代个体。 -
交叉(Crossover):
对选出的父代个体进行交叉操作,通过模拟基因交换过程,生成新的个体。假设选中的两个父代个体分别为个体A和个体B,交叉操作可以通过如下方式进行:子代个体 = [ A [ 1 : k ] , B [ k + 1 : e n d ] ] \text{子代个体} = [A[1:k], B[k+1:end]] 子代个体=[A[1:k],B[k+1:end]]
其中, A [ 1 : k ] A[1:k] A[1:k] 表示个体A的前k个基因, B [ k + 1 : e n d ] B[k+1:end] B[k+1:end] 表示个体B的第k+1个基因到最后一个基因。
-
变异(Mutation):
对交叉得到的子代个体进行变异操作,通过随机改变染色体中的某些基因值来引入新的解。变异操作可以用如下方式实现:子代个体 [ i ] = 子代个体 [ i ] + 随机增量 \text{子代个体}[i] = \text{子代个体}[i] + \text{随机增量} 子代个体[i]=子代个体[i]+随机增量
其中, 子代个体 [ i ] \text{子代个体}[i] 子代个体[i] 表示子代个体中的第i个基因, 随机增量 \text{随机增量} 随机增量 是一个随机数,用于在某个范围内调整基因的值。
-
生成下一代种群:
将交叉和变异得到的子代个体与上一代的个体结合,形成下一代种群。 -
终止条件:
根据预定的终止条件(如迭代次数、达到某个适应度阈值等),判断是否结束优化过程。 -
解码:
将优化后的染色体解码,得到BP神经网络的新参数。 -
更新BP神经网络参数:
将解码得到的新参数应用于BP神经网络,更新其权重和偏置。 -
重复迭代:
重复进行步骤5到步骤12,直到满足终止条件为止。
2.文件结构
goat
arithXover.m
delta.m
ga.m
gabpEval.m
initializega.m
maxGenTerm.m
nonUnifMutation.m
normGeomSelect.m
parse.m
gadecod.m
main.m % 主函数
数据集.xlsx % 可替换数据集
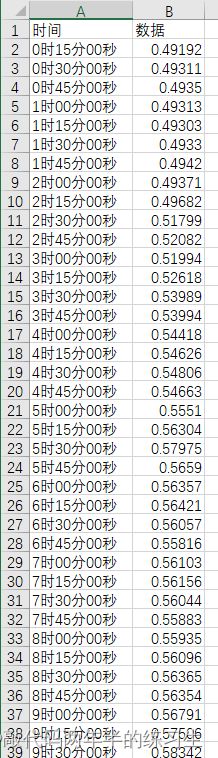
3.Excel数据
4.分块代码
4.1 arithXover.m
function [C1, C2] = arithXover(P1, P2, ~, ~)
%% Arith 交叉采用两个父节点 P1、P2 并沿两个父节点形成的线执行插值。
% P1 - the first parent ( [solution string function value] )
% P2 - the second parent ( [solution string function value] )
% bounds - the bounds matrix for the solution space
% Ops - Options matrix for arith crossover [gen #ArithXovers]
%% 选择一个随机的混合量
a = rand;
%% 创建子代
C1 = P1 * a + P2 * (1 - a);
C2 = P1 * (1 - a) + P2 * a;
4.2 delta.m
function change = delta(ct, mt, y, b)
% delta 函数是非均匀突变使用的非均匀分布。
% 此函数根据当前发电量、最大发电量和可能的偏差量返回变化。
%
% ct - current generation
% mt - maximum generation
% y - maximum amount of change, i.e. distance from parameter value to bounds
% b - shape parameter
%%
r = ct / mt;
if(r > 1)
r = 0.99;
end
change = y * (rand * (1 - r)) ^ b;
4.3 ga.m
function [x, endPop, bPop, traceInfo] = ga(bounds, evalFN, evalOps, startPop, opts, ...
termFN, termOps, selectFN, selectOps, xOverFNs, xOverOps, mutFNs, mutOps)
% Output Arguments:
% x - the best solution found during the course of the run
% endPop - the final population
% bPop - a trace of the best population
% traceInfo - a matrix of best and means of the ga for each generation
%
% Input Arguments:
% bounds - a matrix of upper and lower bounds on the variables
% evalFN - the name of the evaluation .m function
% evalOps - options to pass to the evaluation function ([NULL])
% startPop - a matrix of solutions that can be initialized
% from initialize.m
% opts - [epsilon prob_ops display] change required to consider two
% solutions different, prob_ops 0 if you want to apply the
% genetic operators probabilisticly to each solution, 1 if
% you are supplying a deterministic number of operator
% applications and display is 1 to output progress 0 for
% quiet. ([1e-6 1 0])
% termFN - name of the .m termination function (['maxGenTerm'])
% termOps - options string to be passed to the termination function
% ([100]).
% selectFN - name of the .m selection function (['normGeomSelect'])
% selectOpts - options string to be passed to select after
% select(pop,#,opts) ([0.08])
% xOverFNS - a string containing blank seperated names of Xover.m
% files (['arithXover heuristicXover simpleXover'])
% xOverOps - A matrix of options to pass to Xover.m files with the
% first column being the number of that xOver to perform
% similiarly for mutation ([2 0;2 3;2 0])
% mutFNs - a string containing blank seperated names of mutation.m
% files (['boundaryMutation multiNonUnifMutation ...
% nonUnifMutation unifMutation'])
% mutOps - A matrix of options to pass to Xover.m files with the
% first column being the number of that xOver to perform
% similiarly for mutation ([4 0 0;6 100 3;4 100 3;4 0 0])
%% 初始化参数
n = nargin;
if n < 2 || n == 6 || n == 10 || n == 12
disp('Insufficient arguements')
end
% 默认评估选项
if n < 3
evalOps = [];
end
% 默认参数
if n < 5
opts = [1e-6, 1, 0];
end
% 默认参数
if isempty(opts)
opts = [1e-6, 1, 0];
end
%% 判断是否为m文件
if any(evalFN < 48)
% 浮点数编码
if opts(2) == 1
e1str = ['x=c1; c1(xZomeLength)=', evalFN ';'];
e2str = ['x=c2; c2(xZomeLength)=', evalFN ';'];
% 二进制编码
else
e1str = ['x=b2f(endPop(j,:),bounds,bits); endPop(j,xZomeLength)=', evalFN ';'];
end
else
% 浮点数编码
if opts(2) == 1
e1str = ['[c1 c1(xZomeLength)]=' evalFN '(c1,[gen evalOps]);'];
e2str = ['[c2 c2(xZomeLength)]=' evalFN '(c2,[gen evalOps]);'];
% 二进制编码
else
e1str=['x=b2f(endPop(j,:),bounds,bits);[x v]=' evalFN ...
'(x,[gen evalOps]); endPop(j,:)=[f2b(x,bounds,bits) v];'];
end
end
%% 默认终止信息
if n < 6
termOps = 100;
termFN = 'maxGenTerm';
end
%% 默认变异信息
if n < 12
% 浮点数编码
if opts(2) == 1
mutFNs = 'boundaryMutation multiNonUnifMutation nonUnifMutation unifMutation';
mutOps = [4, 0, 0; 6, termOps(1), 3; 4, termOps(1), 3;4, 0, 0];
% 二进制编码
else
mutFNs = 'binaryMutation';
mutOps = 0.05;
end
end
%% 默认交叉信息
if n < 10
% 浮点数编码
if opts(2) == 1
xOverFNs = 'arithXover heuristicXover simpleXover';
xOverOps = [2, 0; 2, 3; 2, 0];
% 二进制编码
else
xOverFNs = 'simpleXover';
xOverOps = 0.6;
end
end
%% 仅默认选择选项,即轮盘赌。
if n < 9
selectOps = [];
end
%% 默认选择信息
if n < 8
selectFN = 'normGeomSelect';
selectOps = 0.08;
end
%% 默认终止信息
if n < 6
termOps = 100;
termFN = 'maxGenTerm';
end
%% 没有定的初始种群
if n < 4
startPop = [];
end
%% 随机生成种群
if isempty(startPop)
startPop = initializega(80, bounds, evalFN, evalOps, opts(1: 2));
end
%% 二进制编码
if opts(2) == 0
bits = calcbits(bounds, opts(1));
end
%% 参数设置
xOverFNs = parse(xOverFNs);
mutFNs = parse(mutFNs);
xZomeLength = size(startPop, 2); % xzome 的长度
numVar = xZomeLength - 1; % 变量数
popSize = size(startPop,1); % 种群人口个数
endPop = zeros(popSize, xZomeLength); % 第二种群矩阵
numXOvers = size(xOverFNs, 1); % Number of Crossover operators
numMuts = size(mutFNs, 1); % Number of Mutation operators
epsilon = opts(1); % Threshold for two fittness to differ
oval = max(startPop(:, xZomeLength)); % Best value in start pop
bFoundIn = 1; % Number of times best has changed
done = 0; % Done with simulated evolution
gen = 1; % Current Generation Number
collectTrace = (nargout > 3); % Should we collect info every gen
floatGA = opts(2) == 1; % Probabilistic application of ops
display = opts(3); % Display progress
%% 精英模型
while(~done)
[bval, bindx] = max(startPop(:, xZomeLength)); % Best of current pop
best = startPop(bindx, :);
if collectTrace
traceInfo(gen, 1) = gen; % current generation
traceInfo(gen, 2) = startPop(bindx, xZomeLength); % Best fittness
traceInfo(gen, 3) = mean(startPop(:, xZomeLength)); % Avg fittness
traceInfo(gen, 4) = std(startPop(:, xZomeLength));
end
%% 最佳解
if ( (abs(bval - oval) > epsilon) || (gen==1))
% 更新显示
if display
fprintf(1, '\n%d %f\n', gen, bval);
end
% 更新种群矩阵
if floatGA
bPop(bFoundIn, :) = [gen, startPop(bindx, :)];
else
bPop(bFoundIn, :) = [gen, b2f(startPop(bindx, 1 : numVar), bounds, bits)...
startPop(bindx, xZomeLength)];
end
bFoundIn = bFoundIn + 1; % Update number of changes
oval = bval; % Update the best val
else
if display
fprintf(1,'%d ',gen); % Otherwise just update num gen
end
end
%% 选择种群
endPop = feval(selectFN, startPop, [gen, selectOps]);
% 以参数为操作数的模型运行
if floatGA
for i = 1 : numXOvers
for j = 1 : xOverOps(i, 1)
a = round(rand * (popSize - 1) + 1); % Pick a parent
b = round(rand * (popSize - 1) + 1); % Pick another parent
xN = deblank(xOverFNs(i, :)); % Get the name of crossover function
[c1, c2] = feval(xN, endPop(a, :), endPop(b, :), bounds, [gen, xOverOps(i, :)]);
% Make sure we created a new
if c1(1 : numVar) == endPop(a, (1 : numVar))
c1(xZomeLength) = endPop(a, xZomeLength);
elseif c1(1:numVar) == endPop(b, (1 : numVar))
c1(xZomeLength) = endPop(b, xZomeLength);
else
eval(e1str);
end
if c2(1 : numVar) == endPop(a, (1 : numVar))
c2(xZomeLength) = endPop(a, xZomeLength);
elseif c2(1 : numVar) == endPop(b, (1 : numVar))
c2(xZomeLength) = endPop(b, xZomeLength);
else
eval(e2str);
end
endPop(a, :) = c1;
endPop(b, :) = c2;
end
end
for i = 1 : numMuts
for j = 1 : mutOps(i, 1)
a = round(rand * (popSize - 1) + 1);
c1 = feval(deblank(mutFNs(i, :)), endPop(a, :), bounds, [gen, mutOps(i, :)]);
if c1(1 : numVar) == endPop(a, (1 : numVar))
c1(xZomeLength) = endPop(a, xZomeLength);
else
eval(e1str);
end
endPop(a, :) = c1;
end
end
%% 运行遗传算子的概率模型
else
for i = 1 : numXOvers
xN = deblank(xOverFNs(i, :));
cp = find((rand(popSize, 1) < xOverOps(i, 1)) == 1);
if rem(size(cp, 1), 2)
cp = cp(1 : (size(cp, 1) - 1));
end
cp = reshape(cp, size(cp, 1) / 2, 2);
for j = 1 : size(cp, 1)
a = cp(j, 1);
b = cp(j, 2);
[endPop(a, :), endPop(b, :)] = feval(xN, endPop(a, :), endPop(b, :), ...
bounds, [gen, xOverOps(i, :)]);
end
end
for i = 1 : numMuts
mN = deblank(mutFNs(i, :));
for j = 1 : popSize
endPop(j, :) = feval(mN, endPop(j, :), bounds, [gen, mutOps(i, :)]);
eval(e1str);
end
end
end
% 更新记录
gen = gen + 1;
done = feval(termFN, [gen, termOps], bPop, endPop); % See if the ga is done
startPop = endPop; % Swap the populations
[~, bindx] = min(startPop(:, xZomeLength)); % Keep the best solution
startPop(bindx, :) = best; % replace it with the worst
end
[bval, bindx] = max(startPop(:, xZomeLength));
%% 显示结果
if display
fprintf(1, '\n%d %f\n', gen, bval);
end
%% 二进制编码
x = startPop(bindx, :);
if opts(2) == 0
x = b2f(x, bounds,bits);
bPop(bFoundIn, :) = [gen, b2f(startPop(bindx, 1 : numVar), bounds, bits)...
startPop(bindx, xZomeLength)];
else
bPop(bFoundIn, :) = [gen, startPop(bindx, :)];
end
%% 赋值
if collectTrace
traceInfo(gen, 1) = gen; % 当前迭代次数
traceInfo(gen, 2) = startPop(bindx, xZomeLength); % 最佳适应度
traceInfo(gen, 3) = mean(startPop(:, xZomeLength)); % 平均适应度
end
4.4 gabpEval.m
function [sol, val] = gabpEval(sol, ~)
%% 解码适应度值
val = gadecod(sol);
4.5 initializega.m
function pop = initializega(num, bounds, evalFN, evalOps, options)
%% 种群初始化
% initializega creates a matrix of random numbers with
% a number of rows equal to the populationSize and a number
% columns equal to the number of rows in bounds plus 1 for
% the f(x) value which is found by applying the evalFN.
% This is used by the ga to create the population if it
% is not supplied.
%
% pop - the initial, evaluated, random population
% populatoinSize - the size of the population, i.e. the number to create
% variableBounds - a matrix which contains the bounds of each variable, i.e.
% [var1_high var1_low; var2_high var2_low; ....]
% evalFN - the evaluation fn, usually the name of the .m file for
% evaluation
% evalOps - any options to be passed to the eval function defaults []
% options - options to the initialize function, ie.
% [type prec] where eps is the epsilon value
% and the second option is 1 for float and 0 for binary,
% prec is the precision of the variables defaults [1e-6 1]
%% 参数初始化
if nargin < 5
options = [1e-6, 1];
end
if nargin < 4
evalOps = [];
end
%% 编码方式
if any(evalFN < 48) % M文件
if options(2) == 1 % 浮点数编码
estr = ['x=pop(i,1); pop(i,xZomeLength)=', evalFN ';'];
else % 二进制编码
estr = ['x=b2f(pop(i,:),bounds,bits); pop(i,xZomeLength)=', evalFN ';'];
end
else % 非M文件
if options(2) == 1 % 浮点数编码
estr = ['[ pop(i,:) pop(i,xZomeLength)]=' evalFN '(pop(i,:),[0 evalOps]);'];
else % 二进制编码
estr = ['x=b2f(pop(i,:),bounds,bits);[x v]=' evalFN ...
'(x,[0 evalOps]); pop(i,:)=[f2b(x,bounds,bits) v];'];
end
end
%% 参数设置
numVars = size(bounds, 1); % 变量数
rng = (bounds(:, 2) - bounds(:, 1))'; % 可变范围
%% 编码方式
if options(2) == 1 % 二进制编码
xZomeLength = numVars + 1; % 字符串的长度是 numVar + fit
pop = zeros(num, xZomeLength); % 分配新种群
pop(:, 1 : numVars) = (ones(num, 1) * rng) .* (rand(num, numVars)) + ...
(ones(num, 1) * bounds(:, 1)');
else % 浮点数编码
bits = calcbits(bounds, options(1));
pop = round(rand(num, sum(bits) + 1));
end
%% 运行文件
for i = 1 : num
eval(estr);
end
4.6 maxGenTerm.m
function done = maxGenTerm(ops, ~, ~)
% 返回 1,即当达到 maximal_generation 时终止 GA。
%
% ops - a vector of options [current_gen maximum_generation]
% bPop - a matrix of best solutions [generation_found solution_string]
% endPop - the current generation of solutions
%%
currentGen = ops(1);
maxGen = ops(2);
done = currentGen >= maxGen;
4.7 nonUnifMutation.m
function parent = nonUnifMutation(parent, bounds, Ops)
%% 非均匀突变基于非均匀概率分布改变父代的参数之一
% parent - the first parent ( [solution string function value] )
% bounds - the bounds matrix for the solution space
% Ops - Options for nonUnifMutate[gen #NonUnifMutations maxGen b]
%% 相关参数设置
cg = Ops(1); % 当前这一代
mg = Ops(3); % 最大代数
bm = Ops(4); % 形状参数
numVar = size(parent, 2) - 1; % 获取变量个数
mPoint = round(rand * (numVar - 1)) + 1; % 选择一个变量从 1 到变量数随机变化
md = round(rand); % 选择突变方向
if md % 向上限突变
newValue = parent(mPoint) + delta(cg, mg, bounds(mPoint, 2) - parent(mPoint), bm);
else % 向下限突变
newValue = parent(mPoint) - delta(cg, mg, parent(mPoint) - bounds(mPoint, 1), bm);
end
parent(mPoint) = newValue; % 产生子代
4.8 normGeomSelect.m
function newPop = normGeomSelect(oldPop, options)
% NormGeomSelect 是一个基于归一化几何分布的排序选择函数。
% newPop - the new population selected from the oldPop
% oldPop - the current population
% options - options to normGeomSelect [gen probability_of_selecting_best]
%% 交叉选择排序
q = options(2); % 选择最佳的概率
e = size(oldPop, 2); % xZome 的长度,即 numvars + fit
n = size(oldPop, 1); % 种群数目
newPop = zeros(n, e); % 为返回 pop 分配空间
fit = zeros(n, 1); % 为选择概率分配空间
x = zeros(n,2); % rank和id的排序列表
x(:, 1) = (n : -1 : 1)'; % 要知道它是什么元素
[~, x(:, 2)] = sort(oldPop(:, e)); % 排序后获取索引
%% 相关参数
r = q / (1 - (1 - q) ^ n); % 归一化分布,q 素数
fit(x(:, 2)) = r * (1 - q) .^ (x(:, 1) - 1); % 生成选择概率
fit = cumsum(fit); % 计算累积概率
%%
rNums = sort(rand(n, 1)); % 生成 n 个排序的随机数
fitIn = 1; % 初始化循环控制
newIn = 1; % 初始化循环控制
while newIn <= n % 获得 n 个新个体
if(rNums(newIn) < fit(fitIn))
newPop(newIn, :) = oldPop(fitIn, :); % 选择 fitIn 个人
newIn = newIn + 1; % 寻找下一个新人
else
fitIn = fitIn + 1; % 着眼于下一个潜在选择
end
end
4.9 parse.m
function x = parse(inStr)
% parse 是一个函数,它接收一个由空格分隔的文本组成的字符串向量,
% 并将各个字符串项解析为一个 n 项矩阵,每个字符串一行。
% x - the return matrix of strings
% inStr - the blank separated string vector
%% 切割字符串
strLen = size(inStr, 2);
x = blanks(strLen);
wordCount = 1;
last = 0;
for i = 1 : strLen
if inStr(i) == ' '
wordCount = wordCount + 1;
x(wordCount, :) = blanks(strLen);
last = i;
else
x(wordCount, i - last) = inStr(i);
end
end
4.10 gadecod.m
function [val, W1, B1, W2, B2] = gadecod(x)
%% 读取主空间变量
S1 = evalin('base', 'S1'); % 读取隐藏层神经元个数
net = evalin('base', 'net'); % 读取网络参数
p_train = evalin('base', 'p_train'); % 读取输入数据
t_train = evalin('base', 't_train'); % 读取输出数据
%% 参数初始化
R2 = size(p_train, 1); % 输入节点数
S2 = size(t_train, 1); % 输出节点数
%% 输入权重编码
for i = 1 : S1
for k = 1 : R2
W1(i, k) = x(R2 * (i - 1) + k);
end
end
%% 输出权重编码
for i = 1 : S2
for k = 1 : S1
W2(i, k) = x(S1 * (i - 1) + k + R2 * S1);
end
end
%% 隐层偏置编码
for i = 1 : S1
B1(i, 1) = x((R2 * S1 + S1 * S2) + i);
end
%% 输出偏置编码
for i = 1 : S2
B2(i, 1) = x((R2 * S1 + S1 * S2 + S1) + i);
end
%% 赋值并计算
net.IW{1, 1} = W1;
net.LW{2, 1} = W2;
net.b{1} = B1;
net.b{2} = B2;
%% 模型训练
net.trainParam.showWindow = 0; % 关闭训练窗口
net = train(net, p_train, t_train);
%% 仿真测试
t_sim1 = sim(net, p_train);
%% 计算适应度值
val = 1 ./ (sqrt(sum((t_sim1 - t_train).^2) ./ length(t_sim1)));
4.11 main.m
%% 清空环境变量
warning off % 关闭报警信息
close all % 关闭开启的图窗
clear % 清空变量
clc % 清空命令行
%% 导入数据(时间序列的单列数据)
result = xlsread('数据集.xlsx');
%% 添加路径
addpath('goat\')
%% 数据分析
num_samples = length(result); % 样本个数
kim = 15; % 延时步长(kim个历史数据作为自变量)
zim = 1; % 跨zim个时间点进行预测
%% 构造数据集
for i = 1: num_samples - kim - zim + 1
res(i, :) = [reshape(result(i: i + kim - 1), 1, kim), result(i + kim + zim - 1)];
end
%% 划分训练集和测试集
temp = 1: 1: 922;
P_train = res(temp(1: 700), 1: 15)';
T_train = res(temp(1: 700), 16)';
M = size(P_train, 2);
P_test = res(temp(701: end), 1: 15)';
T_test = res(temp(701: end), 16)';
N = size(P_test, 2);
%% 数据归一化
[p_train, ps_input] = mapminmax(P_train, 0, 1);
p_test = mapminmax('apply', P_test, ps_input);
[t_train, ps_output] = mapminmax(T_train, 0, 1);
t_test = mapminmax('apply', T_test, ps_output);
%% 建立模型
S1 = 5; % 隐藏层节点个数
net = newff(p_train, t_train, S1);
%% 设置参数
net.trainParam.epochs = 1000; % 最大迭代次数
net.trainParam.goal = 1e-6; % 设置误差阈值
net.trainParam.lr = 0.01; % 学习率
%% 设置优化参数
gen = 50; % 遗传代数
pop_num = 5; % 种群规模
S = size(p_train, 1) * S1 + S1 * size(t_train, 1) + S1 + size(t_train, 1);
% 优化参数个数
bounds = ones(S, 1) * [-1, 1]; % 优化变量边界
%% 初始化种群
prec = [1e-6, 1]; % epslin 为1e-6, 实数编码
normGeomSelect = 0.09; % 选择函数的参数
arithXover = 2; % 交叉函数的参数
nonUnifMutation = [2 gen 3]; % 变异函数的参数
initPpp = initializega(pop_num, bounds, 'gabpEval', [], prec);
%% 优化算法
[Bestpop, endPop, bPop, trace] = ga(bounds, 'gabpEval', [], initPpp, [prec, 0], 'maxGenTerm', gen,...
'normGeomSelect', normGeomSelect, 'arithXover', arithXover, ...
'nonUnifMutation', nonUnifMutation);
%% 获取最优参数
[val, W1, B1, W2, B2] = gadecod(Bestpop);
%% 参数赋值
net.IW{1, 1} = W1;
net.LW{2, 1} = W2;
net.b{1} = B1;
net.b{2} = B2;
%% 模型训练
net.trainParam.showWindow = 1; % 打开训练窗口
net = train(net, p_train, t_train); % 训练模型
%% 仿真测试
t_sim1 = sim(net, p_train);
t_sim2 = sim(net, p_test );
%% 数据反归一化
T_sim1 = mapminmax('reverse', t_sim1, ps_output);
T_sim2 = mapminmax('reverse', t_sim2, ps_output);
%% 均方根误差
error1 = sqrt(sum((T_sim1 - T_train).^2) ./ M);
error2 = sqrt(sum((T_sim2 - T_test ).^2) ./ N);
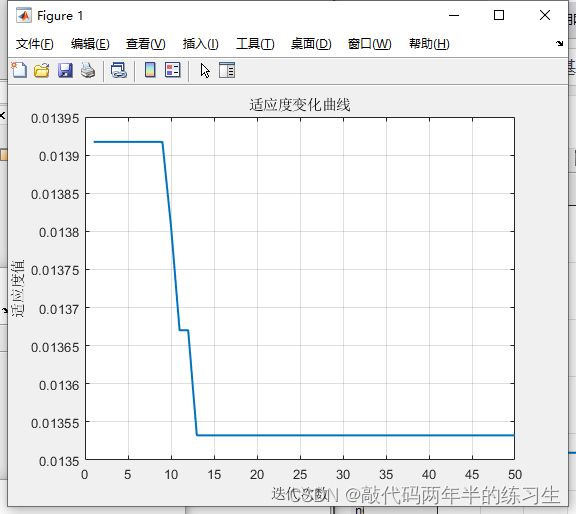
%% 优化迭代曲线
figure
plot(trace(:, 1), 1 ./ trace(:, 2), 'LineWidth', 1.5);
xlabel('迭代次数');
ylabel('适应度值');
string = {'适应度变化曲线'};
title(string)
grid on
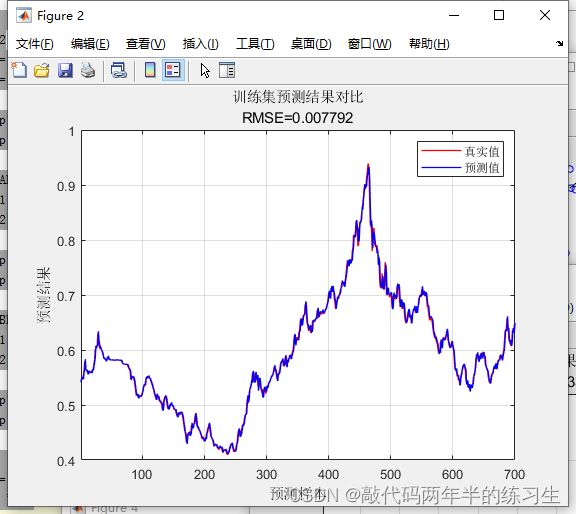
%% 绘图
figure
plot(1: M, T_train, 'r-', 1: M, T_sim1, 'b-', 'LineWidth', 1)
legend('真实值', '预测值')
xlabel('预测样本')
ylabel('预测结果')
string = {'训练集预测结果对比'; ['RMSE=' num2str(error1)]};
title(string)
xlim([1, M])
grid
figure
plot(1: N, T_test, 'r-', 1: N, T_sim2, 'b-', 'LineWidth', 1)
legend('真实值', '预测值')
xlabel('预测样本')
ylabel('预测结果')
string = {'测试集预测结果对比'; ['RMSE=' num2str(error2)]};
title(string)
xlim([1, N])
grid
%% 相关指标计算
% R2
R1 = 1 - norm(T_train - T_sim1)^2 / norm(T_train - mean(T_train))^2;
R2 = 1 - norm(T_test - T_sim2)^2 / norm(T_test - mean(T_test ))^2;
disp(['训练集数据的R2为:', num2str(R1)])
disp(['测试集数据的R2为:', num2str(R2)])
% MAE
mae1 = sum(abs(T_sim1 - T_train)) ./ M ;
mae2 = sum(abs(T_sim2 - T_test )) ./ N ;
disp(['训练集数据的MAE为:', num2str(mae1)])
disp(['测试集数据的MAE为:', num2str(mae2)])
% MBE
mbe1 = sum(T_sim1 - T_train) ./ M ;
mbe2 = sum(T_sim2 - T_test ) ./ N ;
disp(['训练集数据的MBE为:', num2str(mbe1)])
disp(['测试集数据的MBE为:', num2str(mbe2)])
%% 绘制散点图
sz = 25;
c = 'b';
figure
scatter(T_train, T_sim1, sz, c)
hold on
plot(xlim, ylim, '--k')
xlabel('训练集真实值');
ylabel('训练集预测值');
xlim([min(T_train) max(T_train)])
ylim([min(T_sim1) max(T_sim1)])
title('训练集预测值 vs. 训练集真实值')
figure
scatter(T_test, T_sim2, sz, c)
hold on
plot(xlim, ylim, '--k')
xlabel('测试集真实值');
ylabel('测试集预测值');
xlim([min(T_test) max(T_test)])
ylim([min(T_sim2) max(T_sim2)])
title('测试集预测值 vs. 测试集真实值')