信号处理--基于EEG脑电信号的眼睛状态的分析
本实验为生物信息学专题设计小项目。项目目的是通过提供的14导联EEG 脑电信号,实现对于人体睁眼和闭眼两个状态的数据分类分析。每个脑电信号的时长大约为117秒。
目录
加载相关的库函数
读取脑电信号数据并查看数据的属性
绘制脑电多通道连接矩阵
绘制两类数据的相对占比
数据集划分和预处理
模型定义及可视化
模型训练及训练可视化
模型评价
加载相关的库函数
import tensorflow.compat.v1 as tf
from sklearn.metrics import confusion_matrix
import numpy as np
from scipy.io import loadmat
import os
from pywt import wavedec
from functools import reduce
from scipy import signal
from scipy.stats import entropy
from scipy.fft import fft, ifft
import pandas as pd
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.preprocessing import StandardScaler
from tensorflow import keras as K
import matplotlib.pyplot as plt
import scipy
from sklearn import metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
from sklearn.model_selection import KFold,cross_validate
from tensorflow.keras.layers import Dense, Activation, Flatten, concatenate, Input, Dropout, LSTM, Bidirectional,BatchNormalization,PReLU,ReLU,Reshape
from keras.wrappers.scikit_learn import KerasClassifier
from sklearn.metrics import classification_report
from tensorflow.keras.models import Sequential, Model, load_model
import matplotlib.pyplot as plt;
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
from sklearn.decomposition import PCA
from tensorflow import keras
from sklearn.model_selection import cross_val_score
from tensorflow.keras.layers import Conv1D,Conv2D,Add
from tensorflow.keras.layers import MaxPool1D, MaxPooling2D
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')读取脑电信号数据并查看数据的属性
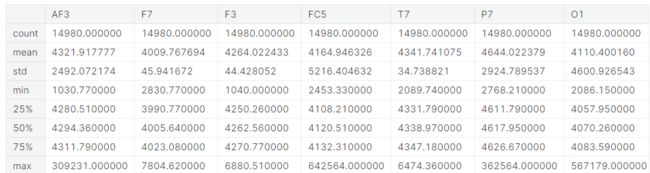
df = pd.read_csv("../input/eye-state-classification-eeg-dataset/EEG_Eye_State_Classification.csv")
df.info()
绘制脑电多通道连接矩阵
plt.figure(figsize = (15,15))
cor_matrix = df.corr()
sns.heatmap(cor_matrix,annot=True)
绘制两类数据的相对占比
# Plotting target distribution
plt.figure(figsize=(6,6))
df['eyeDetection'].value_counts().plot.pie(explode=[0.1,0.1], autopct='%1.1f%%', shadow=True, textprops={'fontsize':16}).set_title("Target distribution")数据集划分和预处理
data = df.copy()
y= data.pop('eyeDetection')
x= data
x_new = StandardScaler().fit_transform(x)
x_new = pd.DataFrame(x_new)
x_new.columns = x.columns
x_train,x_test,y_train,y_test = train_test_split(x_new,y,test_size=0.15)
x_train = np.array(x_train).reshape(-1,14,1)
x_test = np.array(x_test).reshape(-1,14,1)
模型定义及可视化
inputs = tf.keras.Input(shape=(14,1))
Dense1 = Dense(64, activation = 'relu',kernel_regularizer=keras.regularizers.l2())(inputs)
#Dense2 = Dense(128, activation = 'relu',kernel_regularizer=keras.regularizers.l2())(Dense1)
#Dense3 = Dense(256, activation = 'relu',kernel_regularizer=keras.regularizers.l2())(Dense2)
lstm_1= Bidirectional(LSTM(256, return_sequences = True))(Dense1)
drop = Dropout(0.3)(lstm_1)
lstm_3= Bidirectional(LSTM(128, return_sequences = True))(drop)
drop2 = Dropout(0.3)(lstm_3)
flat = Flatten()(drop2)
#Dense_1 = Dense(256, activation = 'relu')(flat)
Dense_2 = Dense(128, activation = 'relu')(flat)
outputs = Dense(1, activation='sigmoid')(Dense_2)
model = tf.keras.Model(inputs, outputs)
model.summary()
tf.keras.utils.plot_model(model)
def train_model(model,x_train, y_train,x_test,y_test, save_to, epoch = 2):
opt_adam = keras.optimizers.Adam(learning_rate=0.001)
es = EarlyStopping(monitor='val_loss', mode='min', verbose=1, patience=10)
mc = ModelCheckpoint(save_to + '_best_model.h5', monitor='val_accuracy', mode='max', verbose=1, save_best_only=True)
lr_schedule = tf.keras.callbacks.LearningRateScheduler(lambda epoch: 0.001 * np.exp(-epoch / 10.))
model.compile(optimizer=opt_adam,
loss=['binary_crossentropy'],
metrics=['accuracy'])
history = model.fit(x_train,y_train,
batch_size=20,
epochs=epoch,
validation_data=(x_test,y_test),
callbacks=[es,mc,lr_schedule])
saved_model = load_model(save_to + '_best_model.h5')
return model,history
模型训练及训练可视化
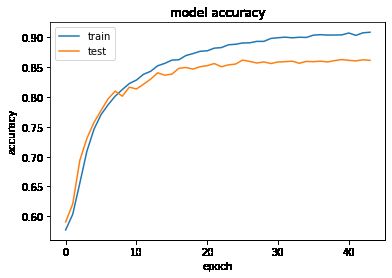
model,history = train_model(model, x_train, y_train,x_test, y_test, save_to= './', epoch = 100)
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
# summarize history for loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
模型评价
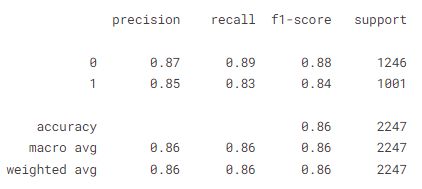
y_pred =model.predict(x_test)
y_pred = np.array(y_pred >= 0.5, dtype = np.int)
confusion_matrix(y_test, y_pred)
print(classification_report(y_test, y_pred))