利用遗传算法解决旅行商问题(GA-TSP)—— Python代码实现
文章目录
- 旅行商问题
- 遗传算法
-
- 编码方案
- 适应度
- 交叉算子
-
- 顺序交叉
- 个体选择
-
-
- 轮盘赌选择
- 锦标赛选择
- 精英选择法
-
- 变异算子
-
- 逆序交叉
- 利用Python编写的GA-TSP算法
-
- 调用函数
- 测试数据
- 可视化
- 算法求解结果
旅行商问题
旅行商问题是指一个旅行商从某个城市出发,依次到达所有城市,使得总路程最短
遗传算法
编码方案
在旅行商问题中,编码方案采取整数编码,即染色体上的每个基因均表示城市的编号,染色体即表示一种旅行商路径。

上述例子中,即表示一种旅行商的路径为7->1->0->4->5->6->3->2
适应度
适应度即表示一个个体对环境的适应能力,适应能力越好的个体应当被保留下来进行繁衍,故适应度越高,对环境的适应能力越高,在每次选择时,应当选择适应度较高的个体进行保留。
在旅行商问题中,每个个体的适应度为路径的总成本大小:
f i t n e s s = d i s t e n c e ( r o u t e ) fitness =distence(route) fitness=distence(route)
以上述个体 7 − 1 − 0 − 4 − 5 − 6 \bold{7-1-0-4-5-6} 7−1−0−4−5−6为例,该个体的适应度即为 f i t n e s s = d i s ( 7 − > 1 ) + d i s ( 1 − > 0 ) + d i s ( 0 − > 4 ) + d i s ( 4 − > 5 ) + d i s ( 4 − > 5 ) + d i s ( 5 − > 6 ) + d i s ( 6 − > 3 ) + d i s ( 3 − > 2 ) + d i s ( 2 − > 7 ) fitness=dis(7->1)+dis(1->0)+dis(0->4)+dis(4->5)+ dis(4->5)+dis(5->6)+dis(6->3)+dis(3->2)+dis(2->7) fitness=dis(7−>1)+dis(1−>0)+dis(0−>4)+dis(4−>5)+dis(4−>5)+dis(5−>6)+dis(6−>3)+dis(3−>2)+dis(2−>7)
即为整条路径的总距离长度
交叉算子
顺序交叉
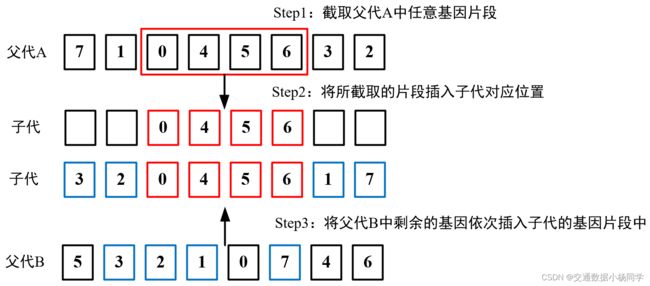
此处采用顺序交叉(Order Crossover, OX)。顺序交叉是一种遗传算法中常用的交叉方法,它用于解决排列问题。在这种交叉方法中,首先从一个父代基因组中随机截取一段基因片段,并将其复制到子代基因组的对应位置上。然后,从另一个父代基因组中选择剩余的基因,按照其在父代基因组中出现的顺序填充到子代基因组中的空位上。这样可以保证子代基因组中基因的顺序与父代基因组相同。
个体选择
轮盘赌选择
轮盘赌选择法(roulette wheel selection)是一种最简单也是最常用的选择方法。在该方法中,各个个体的选择概率和其适应度值成比例,适应度越大,选中概率也越大。轮盘赌选择法的过程如下:
- 计算每个个体的被选中概率 p ( x i ) p(x_i) p(xi)。
- 计算每个部分的累积概率 q ( x i ) q(x_i) q(xi)。
- 随机生成一个数组 m m m,数组中的元素取值范围在0和1之间,并将其按从小到大的方式进行排序。
- 若累积概率 q ( x i ) q(x_i) q(xi)大于数组中的元素 m [ i ] m[i] m[i],则个体 x ( i ) x(i) x(i)被选中,若小于 m [ i ] m[i] m[i],则比较下一个个体 x ( i + 1 ) x(i+1) x(i+1)直至选出一个个体为止。
- 若需要转中 N N N个个体,则将步骤3重复 N N N次即可
锦标赛选择
锦标赛选择法的基本思想是每次从种群中随机取出一定数量的个体,然后选择其中最好的一个进入子代种群。重复该操作,直到新的种群规模达到原来的种群规模。具体的操作步骤如下:
- 确定每次选择的个体数量 N N N(二元锦标赛选择即选择2个个体)。
- 从种群中随机选择 N N N个个体(每个个体被选择的概率相同),根据每个个体的适应度值,选择其中适应度值最好的个体进入下一代种群。
- 重复步骤2多次(重复次数为种群的大小),直到新的种群规模达到原来的种群规模
精英选择法
精英选择法的基本思想是:在每一代中,将种群中适应度最高的若干个体直接复制到下一代种群中,以保留当前种群中的优秀基因。精英选择法可以防止遗传算法在进化过程中出现“早熟”现象,即算法过早地收敛到局部最优解而无法继续搜索全局最优解。精英选择法的步骤如下:
- 在每一代中,计算种群中每个个体的适应度值。
- 从种群中选择适应度值最高的若干个体。
- 将这些个体直接复制到下一代种群中。
- 使用其他选择方法(如轮盘赌选择、锦标赛选择等)从剩余个体中选择其他个体进入下一代种群,直到新的种群规模达到原来的种群规模。
变异算子
逆序交叉
此处采用逆序交叉,首先从个体中随机选择两个位置,然后将这两个位置之间的元素(包括这两个位置)逆序。最后返回变异后的个体。

利用Python编写的GA-TSP算法
class TSP_GA:
def __init__(self,
iteration,
cost_mat,
pop_size,
mutation_rate,
elite_rate,
cross_rate):
self.iteration = iteration # 迭代次数
self.cost_mat = cost_mat # 成本矩阵
self.cities = self.cost_mat.shape[0] # 城市个数
self.pop_size = pop_size # 种群数量
self.mutate_rate = mutation_rate # 变异率
self.elite_rate = elite_rate # 精英率
def initial_population(self): # 种群初始化
population = []
for i in range(self.pop_size):
cities_list = list(range(self.cities))
random.shuffle(cities_list)
population.append(cities_list)
return population
def fitness(self, individual): # 计算每个个体的适应度
route_cost = 0
for i in range(len(individual) - 1):
start = individual[i]
end = individual[i + 1]
route_cost += self.cost_mat[start][end]
route_cost += self.cost_mat[individual[-1]][individual[0]]
return route_cost
@staticmethod
def __crossover(parent_a, parent_b): # 交叉
"""
:param parent_a: 父代基因A
:param parent_b: 父代基因B
:return:
"""
child = [None] * len(parent_a) # 创建一个子代个体
start_index, end_index = random.sample(range(len(parent_a)), 2) # 随机生成两个索引点,用于截取基因片段
if start_index > end_index:
start_index, end_index = end_index, start_index
child[start_index:end_index] = parent_a[start_index:end_index] # 截取父代基因A中的片段,并赋给子代基因的对应位置
remaining_genes = [gene for gene in parent_b if gene not in child] # 从另一个父代基因组中选择剩余的基因
i = 0
for gene in remaining_genes:
while child[i] is not None: # 找到子代基因组中的空位
i += 1
child[i] = gene # 将基因填充到子代基因组中
return child # 返回子代基因组
def select_elites(self, population, fitnesses): # 精英选择
# 计算被选出精英个体的数量
num_elites = int(len(population) * self.elite_rate) # 精英数量
# 根据适应度对种群进行排序
sorted_population = [individual for _, individual in sorted(zip(fitnesses, population))]
# 选取适应度大的前几个
elites = sorted_population[:num_elites]
return elites
def select_two_parents(self, population, fitnesses): #
total_fitness = sum(fitnesses)
selection_probability = [fitness / total_fitness for fitness in fitnesses]
# 选择父代 A
parent_a_index = random.choices(range(len(population)), weights=selection_probability, k=1)[0]
parent_a = population[parent_a_index]
# 选择父代 B
population_without_a = population[:parent_a_index] + population[parent_a_index + 1:]
fitnesses_without_a = fitnesses[:parent_a_index] + fitnesses[parent_a_index + 1:]
total_fitness = sum(fitnesses_without_a)
selection_probability = [fitness / total_fitness for fitness in fitnesses_without_a]
parent_b_index = random.choices(range(len(population_without_a)), weights=selection_probability, k=1)[0]
parent_b = population_without_a[parent_b_index]
return parent_a, parent_b
def displacement_mutation(self, individual):
"""
置换变异
:param individual: 个体
"""
i, j = sorted(random.sample(range(len(individual)), 2))
k = random.randint(0, len(individual) - (j - i + 1))
genes = individual[i:j + 1]
del individual[i:j + 1]
individual[k:k] = genes
return individual
def solve(self):
population = self.initial_population() # init polpulation
best_fitness = []
for i in range(self.iteration): # iteration
fitnesses = [self.fitness(individual) for individual in population] # 求解每个个体的适应度并保存为列表
next_population = self.select_elites(population, fitnesses) # 精英选择
while len(next_population) < self.pop_size:
parent_a, parent_b = self.select_two_parents(population, fitnesses)
child_a = self.__crossover(parent_a, parent_b) # 交叉,生成子代个体a
child_b = self.__crossover(parent_b, parent_a) # 交叉,生成子代个体b
if random.random() < self.mutate_rate:
child_a = self.displacement_mutation(child_a) # 变异
if random.random() < self.mutate_rate:
child_b = self.displacement_mutation(child_b) # 变异
next_population.append(child_a) # 将子代基因组添加到新的种群中
next_population.append(child_b)
population = next_population
fitnesses = [self.fitness(individual) for individual in population] # 求解每个个体的适应度并保存为列表
best_fitness.append(min(fitnesses))
print('当前迭代进度:{}/{},最佳适应度为:{}'.format(i, self.iteration, min(fitnesses)))
fitnesses = [self.fitness(individual) for individual in population]
best_individual = population[fitnesses.index(min(fitnesses))]
return best_individual, min(fitnesses), best_fitness
调用函数
TSP= TSP_GA(teration = 10000,
cost_mat = distence_matrix,
pop_size = 150,
mutation_rate = 0.05,
elite_rate = 0.4,
cross_rate = 0.7) # 实例化,初始化参数
TSP_RESULT = TSP.solve() # 调用TSP_GA中的 solve求解函数
测试数据
| ID | X | Y |
|---|---|---|
| 1 | 1150 | 1760 |
| 2 | 630 | 1660 |
| 3 | 40 | 2090 |
| 4 | 750 | 1100 |
| 5 | 750 | 2030 |
| 6 | 1030 | 2070 |
| 7 | 1650 | 650 |
| 8 | 1490 | 1630 |
| 9 | 790 | 2260 |
| 10 | 710 | 1310 |
| 11 | 840 | 550 |
| 12 | 1170 | 2300 |
| 13 | 970 | 1340 |
| 14 | 510 | 700 |
| 15 | 750 | 900 |
| 16 | 1280 | 1200 |
| 17 | 230 | 590 |
| 18 | 460 | 860 |
| 19 | 1040 | 950 |
| 20 | 590 | 1390 |
| 21 | 830 | 1770 |
| 22 | 490 | 500 |
| 23 | 1840 | 1240 |
| 24 | 1260 | 1500 |
| 25 | 1280 | 790 |
| 26 | 490 | 2130 |
| 27 | 1460 | 1420 |
| 28 | 1260 | 1910 |
| 29 | 360 | 1980 |
| centered | centered | centered |
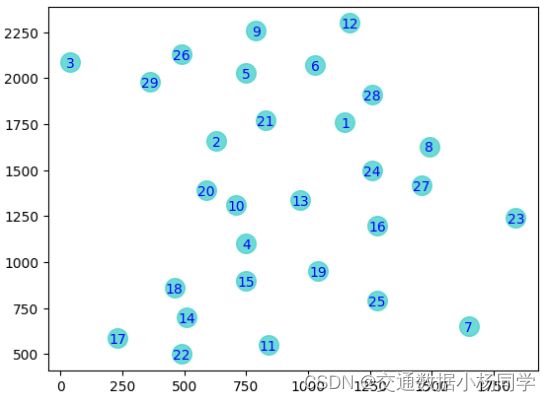
共有29个城市,依次编号为1-29
可视化
data = pd.read_csv('data.csv', header=None) # 测试数据集
data_dic = list(data.itertuples(index=False, name=None))
distances = squareform(pdist(data_dic)) # 生成距离矩阵
for point in data_dic:
label, x, y = point
plt.scatter(x, y,c='darkturquoise',s=220)
plt.text(x, y, label, ha='center', va='center_baseline',fontdict={'size': 10, 'color': 'black'})
算法求解结果

此处由于城市数量过多,所以收敛的可能比较慢,在使用过程中需要根据实际需要调整变异概率、交叉概率以及不同算子,使得每次迭代所得到的种群多样性更丰富,进而使得算法尽快的收敛。