R语言【paleobioDB】——pbdb_richness():绘制指定类群的数量丰度
Package paleobioDB version 0.7.0
paleobioDB 包在2020年已经停止更新,该包依赖PBDB v1 API。
可以选择在Index of /src/contrib/Archive/paleobioDB (r-project.org)下载安装包后,执行本地安装。
Usage
pbdb_richness (data, rank, res, temporal_extent, colour, bord, do.plot)Arguments
参数【data】:输入的数据,数据帧格式。可以通过 pbdb_occurrences() 函数 传参 show = c("phylo", "ident") 获得数据。
参数【rank】:设置感兴趣的分类阶元。可选项包括:“species”,“genus”,“family”,“order”,“class” 和 “phylum”。默认值为 “species”。
参数【temporal_extent】:设置时间范围,向量型(min,max)。
参数【res】:数值型。设置时间范围的时间段刻度。
参数【orig_ext】:1 表示出现,2 表示灭绝。
参数【colour】:改变图中柱子的颜色。默认为 skyblue2。
参数【bord】:设置图形边界的颜色。
参数【do.plot】:TRUE/FALSE。默认为 TRUE。
Value
返回一个数据帧和一个图像,展示了指定类群阶元在特定时间段内和分辨率上的丰富度变化。
Example
library(paleobioDB)
library(RCurl)
options(RCurlOptions = list(cainfo = system.file("CurlSSL", "cacert.pem", package = "RCurl")))
data<- pbdb_occurrences (limit="all", vocab="pbdb",
base_name="Canidae", show=c("phylo", "ident"))
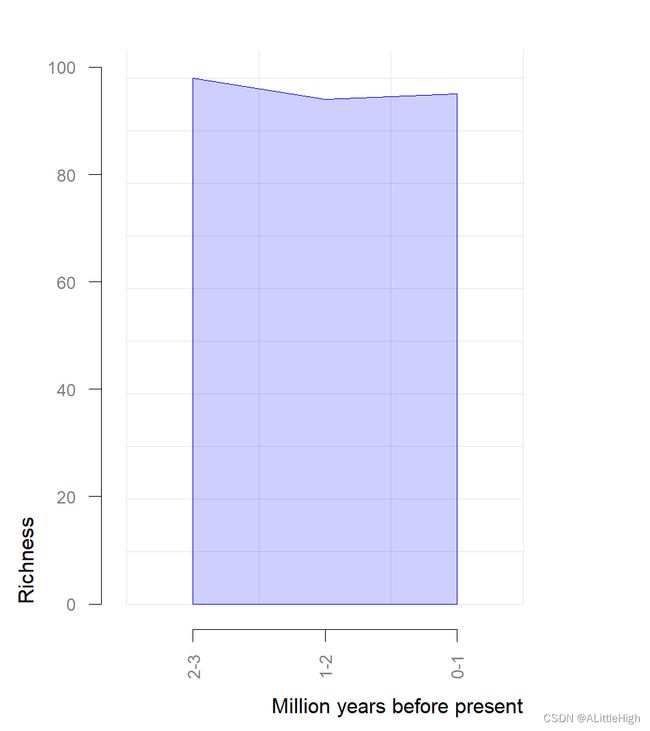
pbdb_richness (data, rank="species", res=1, temporal_extent=c(0,3))
temporal_intervals richness
1 0-1 95
2 1-2 94
3 2-3 98Page
function (data, rank, res = 1, temporal_extent = c(0, 10), colour = "#0000FF30",
bord = "#0000FF", do.plot = TRUE)
{
temporal_range <- pbdb_temp_range(data = data, rank = rank,
do.plot = FALSE)
te <- temporal_extent
time <- seq(from = min(te), to = (max(te)), by = res)
means <- NULL
for (i in 1:length(time) - 1) {
x <- (time[i + 1] + time[i])/2
means <- c(means, x)
}
a <- NULL
for (i in 1:(length(time) - 1)) {
b <- temporal_range[, 1] > time[i] & temporal_range[,
2] <= time[i + 1]
a <- cbind(a, b)
}
richness <- colSums(a + 0, na.rm = T)
temporal_intervals <- paste(time[-length(time)], time[-1],
sep = "-")
richness <- data.frame(temporal_intervals, richness)
if (do.plot == TRUE) {
plot.new()
par(mar = c(5, 5, 1, 5), font.lab = 1, col.lab = "grey20",
col.axis = "grey50", cex.axis = 0.8)
plot.window(xlim = c(max(te), min(te)), xaxs = "i", ylim = c(0,
(max(richness[, 2])) + (max(richness[, 2])/10)),
yaxs = "i")
abline(v = seq(min(te), max(te), by = res), col = "grey90",
lwd = 1)
abline(h = seq(0, max(richness[, 2]) + (max(richness[,
2])/10), by = (max(richness[, 2])/10)), col = "grey90",
lwd = 1)
xx <- c(means[1], means, means[length(means)])
yy <- c(0, richness[, 2], 0)
polygon(xx, yy, col = colour, border = bord)
axis(1, line = 1, las = 2, labels = temporal_intervals,
at = means)
axis(2, line = 1, las = 1)
mtext("Million years before present", line = 3.5, adj = 1,
side = 1)
mtext("Richness", line = 3.5, adj = 0, side = 2)
}
return(richness)
}