学校服务器hpc东南大学,下载国家基因组科技中心数据 gsa-human ascp chatpt建议 Linux系统中写代码
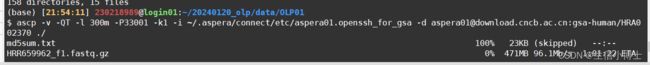
使用ascp批量下载数据
You
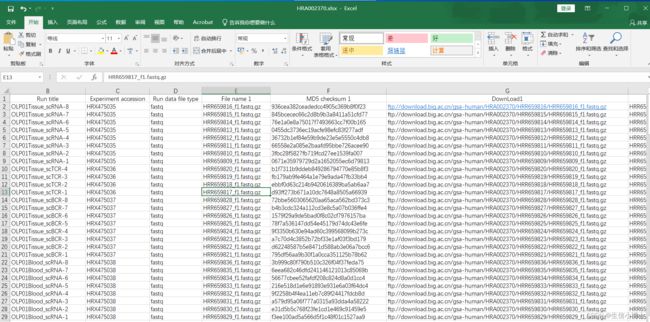
files.csv
帮我写个批量下载的脚本,批量下载时候,把路径中最后的HRR659816批量替换成 Accession列的内容就行了。下面是示例
ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659816 ./-d 参数
1 读取样本Accession
2 python批量生成代码
# Generate the Aspera download commands with replaced Accession numbers
commands = []
base_command = "ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/{accession} ./"
for accession in df['Accession']:
command = base_command.format(accession=accession)
commands.append(command)
# Save the commands to a file
commands_file_path = '/mnt/data/download_commands.sh'
with open(commands_file_path, 'w') as file:
for command in commands:
file.write(command + '\n')
commands_file_path
2.5 同时我想把下载下来的文件,放入文件中各自的sample列的文件夹
# Generate the Aspera download commands with directories for each sample
updated_commands = []
base_command = "mkdir -p {sample_dir} && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/{accession} {sample_dir}/"
for _, row in df.iterrows():
sample_dir = row['sample']
accession = row['Accession']
command = base_command.format(sample_dir=sample_dir, accession=accession)
updated_commands.append(command)
# Save the updated commands to a file
updated_commands_file_path = '/mnt/data/updated_download_commands.sh'
with open(updated_commands_file_path, 'w') as file:
for command in updated_commands:
file.write(command + '\n')
updated_commands_file_path
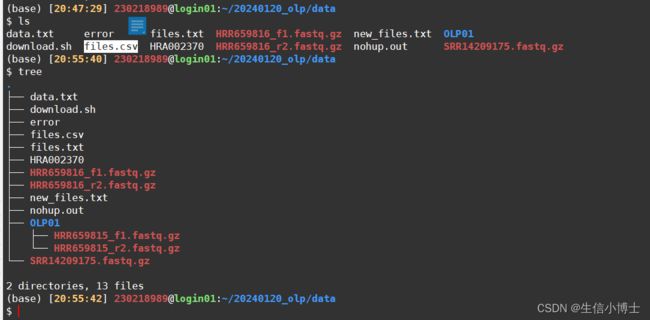
3 最终生成的代码如下
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659815 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659814 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659813 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659812 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659811 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659810 OLP01/
mkdir -p OLP01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659809 OLP01/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659864 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659863 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659862 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659861 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659860 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659859 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659858 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659857 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659856 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659855 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659854 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659853 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659852 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659851 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659850 OLP02/
mkdir -p OLP02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659849 OLP02/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659900 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659899 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659898 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659897 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659896 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659895 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659894 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659893 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659892 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659891 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659890 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659889 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659888 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659887 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659886 OLP03/
mkdir -p OLP03 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659885 OLP03/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659924 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659923 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659922 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659921 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659920 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659919 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659918 OLP04/
mkdir -p OLP04 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659917 OLP04/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659944 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659943 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659942 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659941 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659940 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659939 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659938 Healthy01/
mkdir -p Healthy01 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659937 Healthy01/
mkdir -p Healthy02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659950 Healthy02/
mkdir -p Healthy02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659949 Healthy02/
mkdir -p Healthy02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659948 Healthy02/
mkdir -p Healthy02 && ascp -v -QT -l 300m -P33001 -k1 -i ~/.aspera/connect/etc/aspera01.openssh_for_gsa -d [email protected]:gsa-human/HRA002370/HRR659947 Healthy02/
- Open a terminal and navigate to the directory where you downloaded the script.
- Give the script execution permissions:
chmod +x download_commands.sh - Run the script:
./download_commands.sh
如果使用wget下载数据的话,速度很慢
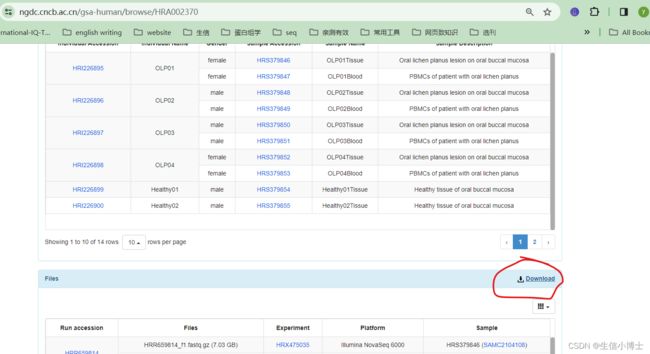
1 首先从下面网址中下载excel
https://ngdc.cncb.ac.cn/gsa-human/browse/HRA002370
https://download.cncb.ac.cn/gsa-human/HRA002370/
2 使用r整理一下,不整理应该也可以
.libPaths(c("/home/data/refdir/Rlib",
"/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2",
"/usr/local/lib/R/library"))
#####安装archr包##别处复制
.libPaths(c("/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2",
"/home/data/t040413/R/yll/usr/local/lib/R/site-library",
"/usr/local/lib/R/library",
"/home/data/refdir/Rlib/"))
.libPaths()
library(Seurat)
library(ggplot2)
library(dplyr)
filepaths=openxlsx::read.xlsx("~/20240120_olp/HRA002370.xlsx",sheet = 4)
filepaths %>%head()
filepaths=filepaths[grep(filepaths$Run.title,pattern="scRNA") ,]
filepaths=filepaths[grep(filepaths$Run.title,pattern="Tissue") ,]
colnames(filepaths)
filepaths$sample=stringr::str_split(filepaths$Run.title,pattern = "Tissue",simplify = TRUE)[,1]
filepaths=filepaths[,c("Accession",'sample', "File.name.1","File.name.2","DownLoad1", "DownLoad2")]
#
#
# getwd()
#
# # 将输出写入文件
# sink("output.txt")
#
# filepaths[-1 ,]
# sink() # 停止输出到文件
#
write.csv(filepaths[-1,],file = "files.csv",row.names = FALSE, quote = FALSE)
# write.table(filepaths[-1,],file = "files.txt",row.names = FALSE,col.names = FALSE,quote = FALSE)
3 学校hpc下载数据,只能使用wget 很奇怪
conda activate screen
screen -S wget
conda activate scanpy
3.5进入python 下载
import pandas as pd
import os
data=pd.read_csv("files.csv")
# 遍历数据,并创建sample文件夹并下载文件
for index, row in data.iterrows():
accession = row['Accession']
sample = row['sample']
file_name_1 = row['File.name.1']
file_name_2 = row['File.name.2']
download_1 = row['DownLoad1']
download_2 = row['DownLoad2']
# 创建sample文件夹
folder_path = f'./{sample}'
if not os.path.exists(folder_path):
os.makedirs(folder_path)
# 下载文件
os.system(f'wget -c -P {folder_path} {download_1}')
os.system(f'wget -c -P {folder_path} {download_2}')
这样就开始,下载了,慢慢等吧