水平基因转移_百度百科/维基百科
- 中文名
- 基因水平转移
- 外文名
- horizontal gene transfer
- 又 称
- 侧向基因转移
- 发现时间
- 1959年
目录
- 1 历史背景
- 2 形成因素
历史背景
形成因素
Horizontal gene transfer (HGT) refers to the transfer of genes between organisms in a manner other than traditional reproduction. Also termed lateral gene transfer (LGT), it contrasts with vertical transfer, the transmission of genes from the parental generation to offspring via sexual or asexual reproduction. HGT has been shown to be an important factor in the evolution of many organisms.[1]
Horizontal gene transfer is the primary reason for bacterial antibiotic resistance,[1][2][3][4][5] and plays an important role in the evolution of bacteria that can degrade novel compounds such as human-created pesticides[6]and in the evolution, maintenance, and transmission of virulence.[7] This horizontal gene transfer often involves temperate bacteriophages and plasmids.[8][9] Genes that are responsible for antibiotic resistance in one species of bacteria can be transferred to another species of bacteria through various mechanisms (e.g., via F-pilus), subsequently arming the antibiotic resistant genes' recipient against antibiotics, which is becoming a medical challenge to deal with.
Most thinking in genetics has focused upon vertical transfer, but there is a growing awareness that horizontal gene transfer is a highly significant phenomenon and among single-celled organisms perhaps the dominant form of genetic transfer.[10][11]
Artificial horizontal gene transfer is a form of genetic engineering.
Contents
[hide]- 1History
- 2Mechanism
- 3Inference
- 4Viruses
- 5Prokaryotes
- 6Eukaryotes
- 7Artificial horizontal gene transfer
- 8Importance in evolution
- 8.1Scientific American article (2000)
- 8.2Genes
- 9See also
- 10Sources and notes
- 11Further reading
History[edit]
Horizontal genetic transfer was first described in Seattle in 1951 in a publication which demonstrated that the transfer of a viral gene intoCorynebacterium diphtheriae created a virulent from a non-virulent strain,[12] also simultaneously solving the riddle of diphtheria (that patients could be infected with the bacteria but not have any symptoms, and then suddenly convert later or never),[13] and giving the first example for the relevance of the lysogenic cycle.[14] Inter-bacterial gene transfer was first described in Japan in a 1959 publication that demonstrated the transfer of antibiotic resistance between different species of bacteria.[15][16] In the mid-1980s, Syvanen[17] predicted that lateral gene transfer existed, had biological significance, and was involved in shaping evolutionary history from the beginning of life on Earth.
As Jain, Rivera and Lake (1999) put it: "Increasingly, studies of genes and genomes are indicating that considerable horizontal transfer has occurred between prokaryotes"[18] (see also Lake and Rivera, 2007).[19] The phenomenon appears to have had some significance for unicellulareukaryotes as well. As Bapteste et al. (2005) observe, "additional evidence suggests that gene transfer might also be an important evolutionary mechanism in protist evolution."[20]
There is some evidence that even higher plants and animals have been affected and this has raised concerns for safety.[21] It has been suggested that lateral gene transfer to humans from bacteria may play a role in cancer.[22]
Richardson and Palmer (2007) state: "Horizontal gene transfer (HGT) has played a major role in bacterial evolution and is fairly common in certain unicellular eukaryotes. However, the prevalence and importance of HGT in the evolution of multicellular eukaryotes remain unclear."[23]
Due to the increasing amount of evidence suggesting the importance of these phenomena for evolution (see below) molecular biologists such as Peter Gogarten have described horizontal gene transfer as "A New Paradigm for Biology".[24]
Some have argued that the process may be a hidden hazard of genetic engineering as it could allow transgenic DNA to spread from species to species.[21]
Mechanism[edit]
There are several mechanisms for horizontal gene transfer:[1][25][26]
- Transformation, the genetic alteration of a cell resulting from the introduction, uptake and expression of foreign genetic material (DNA orRNA).[27] This process is relatively common in bacteria, but less so in eukaryotes.[28] Transformation is often used in laboratories to insert novel genes into bacteria for experiments or for industrial or medical applications. See also molecular biology and biotechnology.
- Transduction, the process in which bacterial DNA is moved from one bacterium to another by a virus (a bacteriophage, or phage).[27]
- Bacterial conjugation, a process that involves the transfer of DNA via a plasmid from a donor cell to a recombinant recipient cell during cell-to-cell contact.[27]
- Gene transfer agents, virus-like elements encoded by the host that are found in the alphaproteobacteria order Rhodobacterales.[29]
A transposon (jumping gene) is a mobile segment of DNA that can sometimes pick up a resistance gene and insert it into a plasmid or chromosome, thereby inducing horizontal gene transfer of antibiotic resistance.[27]
Inference[edit]
Horizontal gene transfer is typically inferred using bioinformatic methods, either by identifying atypical sequence signatures ("parametric" methods) or by identifying strong discrepancies between the evolutionary history of particular sequences compared to that of their hosts.
Viruses[edit]
The virus called Mimivirus infects amoebae. Another virus, called Sputnik, also infects amoebae, but it cannot reproduce unless mimivirus has already infected the same cell.[30] "Sputnik’s genome reveals further insight into its biology. Although 13 of its genes show little similarity to any other known genes, three are closely related to mimivirus and mamavirus genes, perhaps cannibalized by the tiny virus as it packaged up particles sometime in its history. This suggests that the satellite virus could perform horizontal gene transfer between viruses, paralleling the way that bacteriophages ferry genes between bacteria.".[31] Horizontal transfer is also seen between geminiviruses and tobacco plants.[32]
Prokaryotes[edit]
Horizontal gene transfer is common among bacteria, even among very distantly related ones. This process is thought to be a significant cause of increased drug resistance[1][33] when one bacterial cell acquires resistance, and the resistance genes are transferred to other species.[34][35]Transposition and horizontal gene transfer, along with strong natural selective forces have led to multi-drug resistant strains of S. aureus and many other pathogenic bacteria.[27] Horizontal gene transfer also plays a role in the spread of virulence factors, such as exotoxins and exoenzymes, amongst bacteria.[1] A prime example concerning the spread of exotoxins is the adaptive evolution of Shiga toxins in E. coli through horizontal gene transfer via transduction with Shigella species of bacteria.[36] Strategies to combat certain bacterial infections by targeting these specific virulence factors and mobile genetic elements have been proposed.[7] For example, horizontally transferred genetic elements play important roles in the virulence of E. coli, Salmonella, Streptococcus and Clostridium perfringens.[1]
Eukaryotes[edit]
"Sequence comparisons suggest recent horizontal transfer of many genes among diverse species including across the boundaries of phylogenetic'domains'. Thus determining the phylogenetic history of a species can not be done conclusively by determining evolutionary trees for single genes".[37]
- Analysis of DNA sequences suggests that horizontal gene transfer has occurred within eukaryotes from the chloroplast and mitochondrial genomes to the nuclear genome. As stated in the endosymbiotic theory, chloroplasts and mitochondria probably originated as bacterial endosymbionts of a progenitor to the eukaryotic cell.[38]
- Horizontal transfer occurs from bacteria to some fungi, especially the yeast Saccharomyces cerevisiae.[39]
- The adzuki bean beetle has acquired genetic material from its (non-beneficial) endosymbiont Wolbachia.[40] New examples have recently been reported demonstrating that Wolbachia bacteria represent an important potential source of genetic material in arthropods and filarialnematodes.[41]
- Mitochondrial genes moved to parasites of the Rafflesiaceae plant family from their hosts [42][43] and from chloroplasts of a not-yet-identified plant to the mitochondria of the bean Phaseolus.[44]
- Striga hermonthica, a eudicot, has received a gene from sorghum (Sorghum bicolor) to its nuclear genome.[45] The gene is of unknown functionality.
- Pea aphids (Acyrthosiphon pisum) contain multiple genes from fungi.[46][47] Plants, fungi and microorganisms can synthesize carotenoids, but torulene made by pea aphids is the only carotenoid known to be synthesized by an organism in the animal kingdom.[46]
- The malaria pathogen Plasmodium vivax acquired genetic material from humans that might help facilitate its long stay in the body.[48]
- A bacteriophage-mediated mechanism transfers genes between prokaryotes and eukaryotes. Nuclear localization signals in bacteriophage terminal proteins (TP) prime DNA replication and become covalently linked to the viral genome. The role of virus and bacteriophages in HGT in bacteria, suggests that TP-containing genomes could be a vehicle of inter-kingdom genetic information transference all throughout evolution.[49]
- HhMAN1 is a gene in the genome of the coffee borer beetle (Hypothenemus hampei) that resembles bacterial genes, and is thought to be transferred from bacteria in the beetle's gut.[50][51]
- A gene that allowed ferns to survive in dark forests came from the hornwort, which grows in mats on streambanks or trees. The neochrome gene arrived about 180 million years ago.[52]
- Plants are capable of receiving genetic information from viruses by horizontal gene transfer.[32]
- One study identified approximately 100 of humans' approximately 20,000 total genes which likely resulted from horizontal gene transfer,[53] but this number has been challenged by several researchers arguing these candidate genes for HGT are more likely the result of gene loss combined with differences in the rate of evolution [54]
Update: Genome Biol. 2015 Mar 13;16(1):50. doi: 10.1186/s13059-015-0607-3. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Crisp A, Boschetti C, Perry M, Tunnacliffe A, Micklem G. http://www.ncbi.nlm.nih.gov/pubmed/25785303
Artificial horizontal gene transfer[edit]
Genetic engineering is essentially horizontal gene transfer, albeit with synthetic expression cassettes. TheSleeping Beauty transposon system[55] (SB) was developed as a synthetic gene transfer agent that was based on the known abilities of Tc1/mariner transposons to invade genomes of extremely diverse species.[56] The SB system has been used to introduce genetic sequences into a wide variety of animal genomes.[57][58]
Importance in evolution[edit]
Horizontal gene transfer is a potential confounding factor in inferring phylogenetic trees based on the sequenceof one gene.[59] For example, given two distantly related bacteria that have exchanged a gene a phylogenetic treeincluding those species will show them to be closely related because that gene is the same even though most other genes are dissimilar. For this reason it is often ideal to use other information to infer robust phylogenies such as the presence or absence of genes or, more commonly, to include as wide a range of genes for phylogenetic analysis as possible.
For example, the most common gene to be used for constructing phylogenetic relationships in prokaryotes is the16s rRNA gene since its sequences tend to be conserved among members with close phylogenetic distances, but variable enough that differences can be measured. However, in recent years it has also been argued that 16s rRNA genes can also be horizontally transferred. Although this may be infrequent the validity of 16s rRNA-constructed phylogenetic trees must be reevaluated.[60]
Biologist Johann Peter Gogarten suggests "the original metaphor of a tree no longer fits the data from recent genome research" therefore "biologists should use the metaphor of a mosaic to describe the different histories combined in individual genomes and use the metaphor of a net to visualize the rich exchange and cooperative effects of HGT among microbes."[24] There exist several methods to infer such phylogenetic networks.
Using single genes as phylogenetic markers, it is difficult to trace organismal phylogeny in the presence of horizontal gene transfer. Combining the simple coalescence model of cladogenesis with rare HGT horizontal gene transfer events suggest there was no single most recent common ancestor that contained all of the genes ancestral to those shared among the three domains of life. Each contemporary molecule has its own history and traces back to an individual molecule cenancestor. However, these molecular ancestors were likely to be present in different organisms at different times."[61]
Scientific American article (2000)[edit]
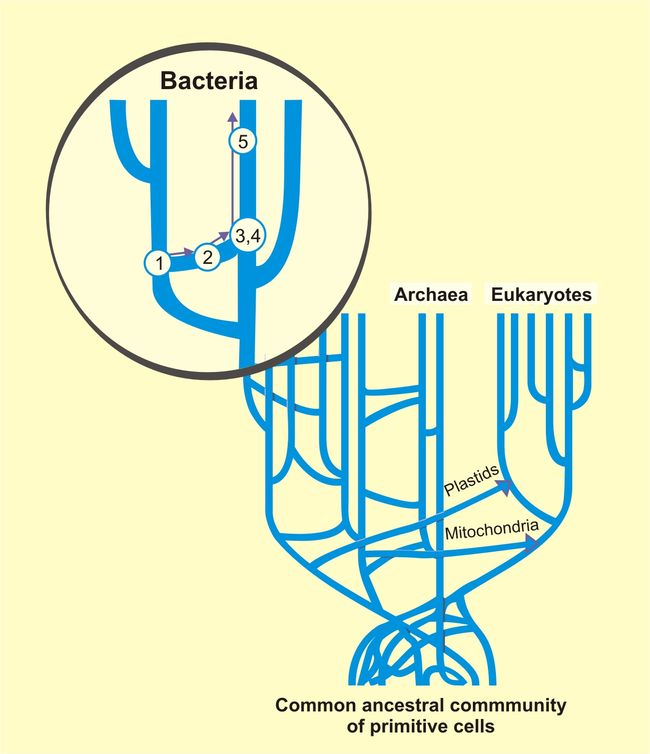
Uprooting the Tree of Life by W. Ford Doolittle (Scientific American, February 2000, pp 90–95)[62] contains a discussion of the Last Universal Common Ancestor and the problems that arose with respect to that concept when one considers horizontal gene transfer. The article covers a wide area — the endosymbiont hypothesis for eukaryotes, the use of small subunit ribosomal RNA (SSU rRNA) as a measure of evolutionary distances (this was the field Carl Woese worked in when formulating the first modern "tree of life", and his research results with SSU rRNA led him to propose theArchaea as a third domain of life) and other relevant topics. Indeed, it was while examining the new three-domain view of life that horizontal gene transfer arose as a complicating issue: Archaeoglobus fulgidus is cited in the article (p. 76) as being an anomaly with respect to a phylogenetictree based upon the encoding for the enzyme HMGCoA reductase — the organism in question is a definite Archaean, with all the cell lipids and transcription machinery that are expected of an Archaean, but whose HMGCoA genes are actually of bacterial origin.[62]
Again on p. 76, the article continues with:
- "The weight of evidence still supports the likelihood that mitochondria in eukaryotes derived from alpha-proteobacterial cells and that chloroplasts came from ingested cyanobacteria, but it is no longer safe to assume that those were the only lateral gene transfers that occurred after the first eukaryotes arose. Only in later, multicellular eukaryotes do we know of definite restrictions on horizontal gene exchange, such as the advent of separated (and protected) germ cells." [62]
The article continues with:
- "If there had never been any lateral gene transfer, all these individual gene trees would have the same topology (the same branching order), and the ancestral genes at the root of each tree would have all been present in the last universal common ancestor, a single ancient cell. But extensive transfer means that neither is the case: gene trees will differ (although many will have regions of similar topology) and there would never have been a single cell that could be called the last universal common ancestor. [62]
- "As Woese has written, 'the ancestor cannot have been a particular organism, a single organismal lineage. It was communal, a loosely knit, diverse conglomeration of primitive cells that evolved as a unit, and it eventually developed to a stage where it broke into several distinct communities, which in their turn became the three primary lines of descent (bacteria, archaea and eukaryotes)' In other words, early cells, each having relatively few genes, differed in many ways. By swapping genes freely, they shared various of their talents with their contemporaries. Eventually this collection of eclectic and changeable cells coalesced into the three basic domains known today. These domains become recognisable because much (though by no means all) of the gene transfer that occurs these days goes on within domains." [62]
With regard to how horizontal gene transfer affects evolutionary theory (common descent, universal phylogenetic tree) Carl Woese says:
- "What elevated common descent to doctrinal status almost certainly was the much later discovery of the universality of biochemistry, which was seemingly impossible to explain otherwise. But that was before horizontal gene transfer (HGT), which could offer an alternative explanation for the universality of biochemistry, was recognized as a major part of the evolutionary dynamic. In questioning the doctrine of common descent, one necessarily questions the universal phylogenetic tree. That compelling tree image resides deep in our representation of biology. But the tree is no more than a graphical device; it is not some a priori form that nature imposes upon the evolutionary process. It is not a matter of whether your data are consistent with a tree, but whether tree topology is a useful way to represent your data. Ordinarily it is, of course, but the universal tree is no ordinary tree, and its root no ordinary root. Under conditions of extreme HGT, there is no (organismal) "tree." Evolution is basically reticulate." [63]
In a May 2010 article in Nature, Douglas Theobald[64] argued that there was indeed one Last Universal Common Ancestor to all existing life and that horizontal gene transfer has not destroyed our ability to infer this.
Genes[edit]
-
This list is incomplete; you can help by expanding it.
There is evidence for historical horizontal transfer of the following genes:
- Lycopene cyclase
- TetO gen conferring resistance to tetracycline, between Campylobacter jejuni.[66]
- Neochrome, gene in some ferns that enhances their ability to survive in dim light. Believed to have been acquired from algae sometime during the Cretaceous.[67]