Coursera NG 机器学习 第七周 KMeans PCA 图像压缩 Python实现
KMeans
ex7.py
import numpy as np
import matplotlib.pyplot as plt
import time
from scipy.io import loadmat
from sklearn.cluster import KMeans
from ex7modules import *
#Part 1:Check MyKMeans
X=loadmat('ex7data2.mat')['X']
K=3
max_iters=10
init_centroids=np.array([[3,3],[6,2],[8,5]])
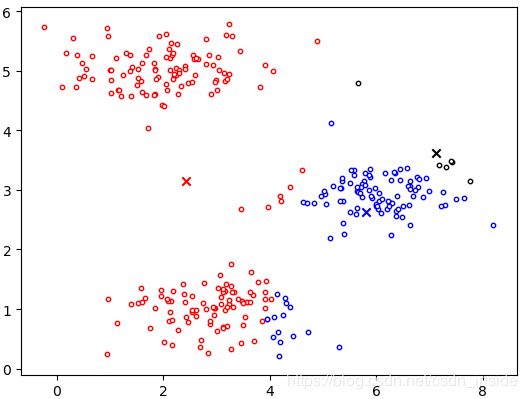
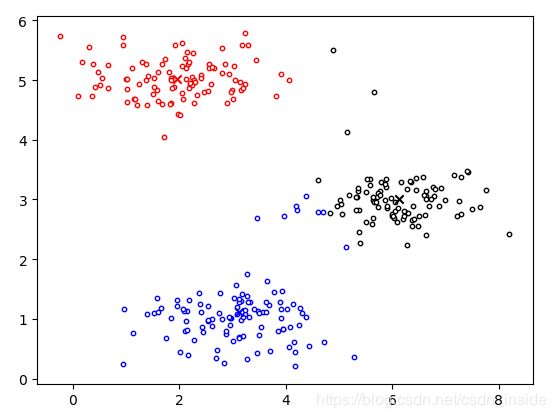
centroids,idx=MyKMeans(X,init_centroids,max_iters,True)
#Part 2:Image Compression
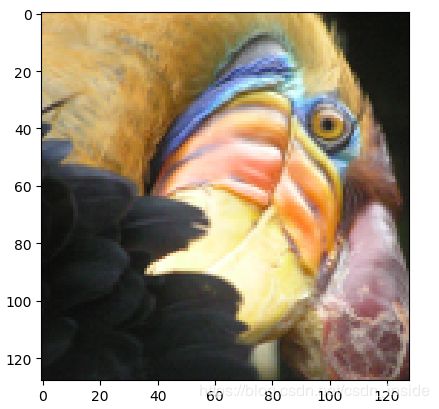
fig=loadmat('bird_small')['A']/255 #Normalize
fig_size=fig.shape[0]
plt.imshow(fig)
plt.show()
fig=fig.reshape(3,-1).T #convert(128,128,3) to (3,128*128) ,to fit the KMeans function
K=16 #reduce nmuber of colors to 16
max_iters=10
time_start=time.time()
init_centroids=InitCentroids(fig,K)
fig_centroids,_=MyKMeans(fig,init_centroids,max_iters,False) # find the most used K colors
fig_idx=findClosestCentroids(fig,fig_centroids) # find every pixel's closest color
time_end=time.time()
print("Using my Kmeans costs time: ",time_end-time_start)
fig_recovered=np.zeros((fig_size*fig_size,3)) #assign every pixel to the closest color
for i in range(fig_size*fig_size):
fig_recovered[i,:]=fig_centroids[fig_idx[i]-1,:]
fig_recovered=fig_recovered.T.reshape((fig_size,fig_size,3)) #need to Transpose first,otherwise
#there is a mistake in image show
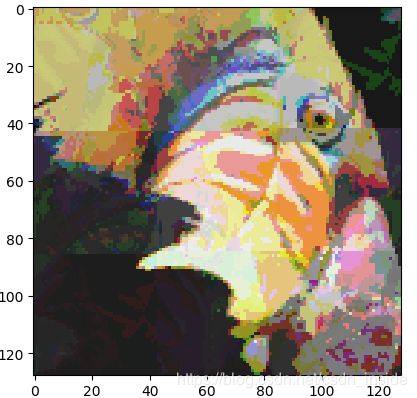
plt.imshow(fig_recovered)
plt.show()
#Part 3:Using SKlearn
time_start=time.time()
clf=KMeans(n_clusters=16,init='random',max_iter=50)
clf.fit(fig)
time_end=time.time()
print("Using SKlearn Kmeans costs time: ",time_end-time_start)
cluster_centers=clf.cluster_centers_
labels=clf.labels_
fig_recovered=np.zeros((fig_size*fig_size,3))
for i in range(fig_size*fig_size):
fig_recovered[i,:]=cluster_centers[labels[i],:]
fig_recovered=fig_recovered.T.reshape((fig_size,fig_size,3))
plt.imshow(fig_recovered)
plt.show()
从左至右,依次是原图,自己的KMeans和SKlearn的KMeans。速度上,还是差很多滴~~~
怎么感觉图被压胡了。。w(゜Д゜)w。。。没有课件里的效果好。。
课件效果
PCA
ex7pca.py
from scipy.io import loadmat
from ex7modules import *
#part 1
X=loadmat('ex7data1.mat')['X']
X_norm,mu,sigma=featureNormalize(X)
U,S=PCA(X_norm)
visualizeEigVector(mu,U,S,X)
print('Top eigenvector: ')
print(U[0,0],U[0,1])
print()
K=1
Z=projectData(X_norm,U,K)
X_rec=recoverData(Z,U,K)
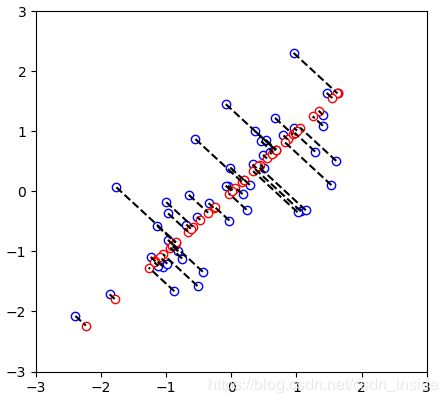
visualizePCA(X_norm,X_rec)
#Part 2
X=loadmat('ex7faces.mat')['X']
displayData(X,10)
X_norm,_,_=featureNormalize(X)
U,S=PCA(X_norm)
displayData(U.T,6)
K=100
Z=projectData(X_norm,U,K)
print('The projected data Z has a size of: ',Z.shape)
X_rec=recoverData(Z,U,K)
displayData(X_rec,10)
Cpcompare(X,X_rec,10)
前100个图像
提取的前36个特征(鬼出没w(゜Д゜)w)
压缩后还原的图像
压缩前跟压缩后的对比
ex7modules.py
import numpy as np
import matplotlib.pyplot as plt
def findClosestCentroids(X,centroids):
K=centroids.shape[0]
dist=np.zeros((X.shape[0],K))
for i in range(X.shape[0]):
for j in range(K):
dist[i, j]=np.linalg.norm(X[i,:]-centroids[j,:])
idx=np.argmin(dist,axis=1)
return idx+1
def computeCentroids(X,idx,K):
centroids=np.zeros((K,X.shape[1]))
for i in range(1,K+1):
X_idx = np.where(idx == i)
X_re = X[X_idx, :].reshape(X[X_idx, :].shape[1], X[X_idx, :].shape[2])
centroids[i - 1, :] = np.sum(X_re, axis=0, keepdims=True) / len(X_idx[0])
return centroids
def MyKMeans(X,initial_centroids,max_iters,plot):
K=initial_centroids.shape[0]
for i in range(max_iters):
idx=findClosestCentroids(X,initial_centroids)
initial_centroids=computeCentroids(X,idx,K)
if plot==True:
idx1 = np.where(idx == 1)[0]
idx2 = np.where(idx == 2)[0]
idx3 = np.where(idx == 3)[0]
plt.scatter(X[idx1, 0], X[idx1, 1], c='w',edgecolors='r',s=10)
plt.scatter(X[idx2, 0], X[idx2, 1], c='w',edgecolors='b',s=10)
plt.scatter(X[idx3, 0], X[idx3, 1], c='w',edgecolors='k',s=10)
plt.scatter(initial_centroids[0, 0], initial_centroids[0, 1], marker='x', c='r')
plt.scatter(initial_centroids[1, 0], initial_centroids[1, 1], marker='x', c='b')
plt.scatter(initial_centroids[2, 0], initial_centroids[2, 1], marker='x', c='k')
plt.show()
return initial_centroids,idx
def InitCentroids(X,K):
randidx=np.random.permutation(X.shape[0])
centroids=X[randidx[0:K],:]
return centroids
def featureNormalize(X):
mu=np.mean(X,axis=0)
sigma=np.std(X,axis=0,ddof=1)
X=(X-mu)/sigma
return X,mu,sigma
def PCA(X):
U,S,_=np.linalg.svd(X.T.dot(X)/X.shape[0])
return U,S
def projectData(X,U,K):
return X.dot(U[:,:K])
def recoverData(Z,U,K):
return Z.dot(U[:,:K].T)
def displayData(X,num):
fig, ax = plt.subplots(num, num)
for i in range(num):
for j in range(num):
ax[i, j].imshow(X[i * num + j, :].reshape((32, 32)).T, cmap='gray')
ax[i, j].set_xticks([])
ax[i, j].set_yticks([])
fig.tight_layout()
plt.subplots_adjust(wspace=0, hspace=0)
plt.show()
def Cpcompare(X,X_rec,num):
fig, ax = plt.subplots(num, num*2)
for i in range(num):
for j in range(0,num*2,2):
ax[i, j].imshow(X[i * num + j, :].reshape((32, 32)).T, cmap='gray')
ax[i, j].set_xticks([])
ax[i, j].set_yticks([])
ax[i, j+1].imshow(X_rec[i * num + j, :].reshape((32, 32)).T, cmap='gray')
ax[i, j+1].set_xticks([])
ax[i, j+1].set_yticks([])
fig.tight_layout()
plt.subplots_adjust(wspace=0, hspace=0)
plt.show()
def visualizeEigVector(mu,U,S,X):
mu = mu.reshape((1, 2))
point1 = mu + 1.5 * S[0] * (U[:, 0].reshape((1, 2)))
point2 = mu + 1.5 * S[1] * (U[:, 1].reshape((1, 2)))
x1 = mu[:, 0]
y1 = mu[:, 1]
x2 = point1[:, 0]
y2 = point1[:, 1]
x3 = point2[:, 0]
y3 = point2[:, 1]
ax1 = np.array([x1, x2])
ax2 = np.array([x1, x3])
ay1 = np.array([y1, y2])
ay2 = np.array([y1, y3])
plt.scatter(X[:, 0], X[:, 1], marker='o', c='w', edgecolors='b')
plt.plot(ax1, ay1, c='k')
plt.plot(ax2, ay2, c='k')
plt.show()
def visualizePCA(X_norm,X_rec):
plt.xlim((-3, 3))
plt.ylim((-3, 3))
plt.scatter(X_norm[:, 0], X_norm[:, 1], marker='o', c='w', edgecolors='b')
plt.scatter(X_rec[:, 0], X_rec[:, 1], marker='o', c='w', edgecolors='r')
for i in range(X_norm.shape[0]):
x = np.array([X_norm[i, 0], X_rec[i, 0]])
y = np.array([X_norm[i, 1], X_rec[i, 1]])
plt.plot(x, y, 'k--')
plt.show()