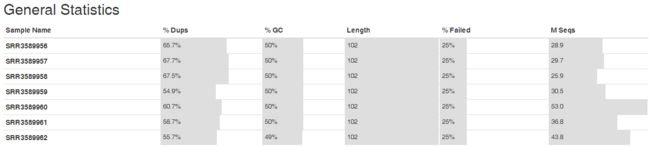
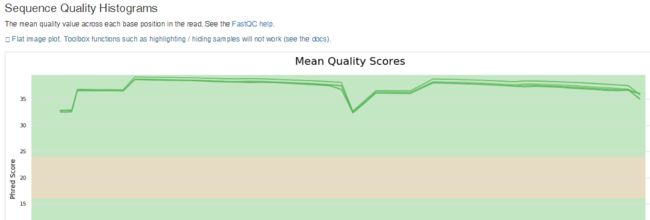
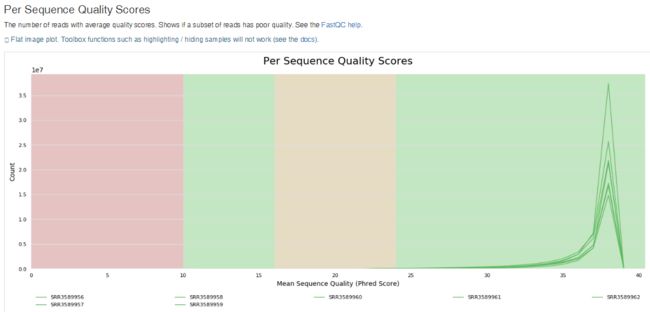
- TCP 三次握手与四次挥手
FHKHH
tcp/ip网络服务器
TCP三次握手与四次挥手知识总结一、TCP连接与断开的核心机制1.三次握手(建立连接)目的:建立客户端与服务端之间的双向传输通道,确保双方都能确认对方的接收和发送能力,为后续的数据传输奠定可靠基础。流程:客户端发送SYN客户端发送SYN报文,请求建立连接,并包含初始序列号(SEQ),此时客户端进入SYN_SENT状态。服务端回应SYN-ACK服务端收到SYN后,回应SYN-ACK,其中ACK为客户
- JavaSE : 注解 Annotation
Edenyt
java-eejava
注解Java中的注解(Annotation)是一种元数据形式,用于向编译器或JVM提供有关程序元素(如类、方法、变量、参数和包)的附加信息。注解不会直接影响程序的行为或结构,但它们可以被编译器、开发工具或运行时环境用于生成代码、进行验证、执行处理或提供信息。以下是关于Java注解的几个关键点:1.注解的种类1.1.内置标准注解:@Override:指示一个方法覆盖了超类中的方法。@Deprecat
- 自然语言处理系列(5)——情感分析的原理与实战
DoYangTan
自然语言处理人工智能
自然语言处理系列(5)——情感分析的原理与实战情感分析(SentimentAnalysis)是自然语言处理中的一项经典任务,目的是通过分析文本,判断其表达的情感倾向性。情感分析广泛应用于社交媒体监控、市场调研、客户服务等领域,帮助企业和机构快速了解用户的情感态度。在本文中,我们将深入探讨情感分析的基本概念、常用方法,并展示如何使用Python和现代NLP工具实现情感分析任务。1.情感分析的基本概念
- 一文读懂西门子 PLC 串口转以太网系列模块
天津三格电子
网络
在工业自动化领域,随着智能化和信息化的不断发展,设备之间的高效通信变得至关重要。西门子PLC作为工业控制的核心设备,其通信方式的拓展需求日益凸显。西门子PLC串口转网口产品应运而生,它为实现串口设备与以太网网络的无缝连接提供了可靠的解决方案。新品上市,欢迎详询1.1产品功能产品可以用来给西门子S7-200/300PLC串口扩展出网口来,扩展出来的网口支持西门子S7TCP协议和ModbusTCP协议
- C进阶 自定义类型
一只自律的鸡
C进阶c语言开发语言
目录前言一结构体二结构体的存储三位段四枚举五联合体总结前言我们之前学习的intchardouble......都是内置类型,但是我们今天所学习的是自定义类型,比如联合体,结构体,枚举一结构体结构体是一些值的集合,这些值统称为成员变量,每个成员都是可以用不同的的基本数据类型结构体的使用场景:结构体的意义在于可以进行封装一个整体的所有变量,这个是十分便捷的,这样就可以不用重复的操作进行重复的定义相同的
- 七个合法学习黑客技术的平台,让你从萌新成为大佬
黑客白帽子黑爷
学习php开发语言web安全网络
1、HackThisSite提供在线IRC聊天和论坛,让用户交流更加方便。网站涵盖多种主题,包括密码破解、网络侦察、漏洞利用、社会工程学等。非常适用于个人提高网络安全技能2、HackaDay涵盖多个领域,包括黑客技术、科技、工程和DIY等内容,站内提供大量有趣的文章、视频、教程和新闻,帮助用户掌握黑客技术和DIY精神。3、OffensiveSecurity一个专门提供网络安全培训和认证的公司,课程
- C 语言文件操作详解
15Moonlight
c语言开发语言
目录1.什么是文件1.1文件名1.2程序文件1.3数据文件2.文件的打开和关闭2.1流和标准流2.1.1流2.1.2标准流2.2文件指针2.3文件的打开和关闭3.文件的顺序读写3.1顺序读写函数3.2使用示例3.2.1fgetc和fputc3.2.2fgets和fputs3.2.3fscanf和fprintf3.2.4fread和fwrite3.3功能对比3.4scanf/fscanf/sscan
- 第十一章:服务器信道管理模块
转调
仿Rabbit消息队列c++消息队列
目录第一节:模块介绍第二节:通信协议第三节:信道模块实现3-1.类型别名定义3-2.Channel类3-3.ChannelManager类下期预告:该模块在mqserver目录下实现。第一节:模块介绍服务器信道的作用是处理来自于客户端的各种请求,然后返回一个响应,那么客户端都有哪些请求呢?比如:交换机的声明与创建、队列的声明与创建、绑定与解绑等。请求的种类如此多,信道要怎么识别这些请求,执行对应的
- 【MySQL | 四、 表的基本查询(增删查改)】
ヾ慈城
mysqlandroidadb
目录表的增删查改Create(创建)表数据的插入替换Retrieve(读取)1.全列查询2.指定列查询3.表达式查询4.为查询结果指定别名5.去重查询WHERE条件查询排序筛选分页查询Update(更新)Delete(删除)删除整张表数据插入查询结果聚合函数groupbyhaving和where的区别1.作用范围不同2.执行顺序不同查询语句执行顺序分析1.**`SELECT`查询语句**2.**`
- SQL笔记#数据更新
月吟荧静
SQL笔记sql笔记数据库
一、数据的插入(INSERT语句的使用方法)1、什么是INSERT首先通过CREATETABLE语句创建表,但创建的表中没有数据;再通过INSERT语句向表中插入数据。--创建表ProductInsCREATETABLEProductIns(product_idCHAR(4)NOTNULL,product_nameVARCHAR(100)NOTNULL,product_typeVARCHAR(32
- 【2025年】全国CTF夺旗赛-从零基础入门到竞赛,看这一篇就稳了!
白帽黑客鹏哥
web安全CTF网络安全大赛pythonLinux
基于入门网络安全/黑客打造的:黑客&网络安全入门&进阶学习资源包目录一、CTF简介二、CTF竞赛模式三、CTF各大题型简介四、CTF学习路线4.1、初期1、html+css+js(2-3天)2、apache+php(4-5天)3、mysql(2-3天)4、python(2-3天)5、burpsuite(1-2天)4.2、中期1、SQL注入(7-8天)2、文件上传(7-8天)3、其他漏洞(14-15
- Java中字符流和字节流的区别
刘小炮吖i
Java后端开发面试题Javajava开发语言
相同点在Java的I/O体系中,字节流和字符流都配备了缓冲机制的实现类,以此显著提升数据读写的效率。字符流:借助BufferedReader和BufferedWriter,它们在处理字符数据时,会将数据先缓存起来,减少与底层数据源或目标的交互次数,从而加速操作。例如,当逐行读取大文本文件时,BufferedReader的缓冲功能能避免频繁的磁盘I/O操作。字节流:BufferedInputStre
- SpringBoot备份神通数据库
松岛的枫叶
springboot数据库后端
SpringBoot备份神通数据库····直接上代码···publicStringbackupOsrdb(){//构建命令Listcommand=newArrayList<>();command.add("osrexp");command.add("-u");command.add("SYSDBA"+"/"+"szoscar55");//用户名/密码command.add("-d");comman
- SQLite自增列相关内容
秦时明月之君临天下
SQLitesqlitejvm数据库
文章目录相关知识创建表插入数据查看自增列重置自增列注意事项参考文档相关知识SQLite的自增用法和MySQL类似:使用的关键字是AUTOINCREMENT(MySQL用的是AUTO_INCREMENT)。AUTOINCREMENT关键字会增加额外的CPU、内存、磁盘空间和磁盘I/O开销,如果不是严格需要的话,应该避免使用。它通常是不需要的。在SQLite中,具有INTEGERPRIMARYKEY类
- Mysql疑难报错排查 - Field ‘XXX‘ doesn‘t have a default value
m0_74823408
面试学习路线阿里巴巴mysql数据库
项目场景:数据库环境:mysql8;工程使用:MyBatisPlus表情况:问题描述某一个插入语句使用了MyBatisPlus的save方法,因为end_time1end_time2都并没有值,所以在MyBatisPlus默认情况下,并不会在插入语句中提及,最终提取其SQL:INSERTINTOaaaa(serial_no,business_date,market_no,report_code)V
- hivePB级迁移方案
我要用代码向我喜欢的女孩表白
数据库bigdata-大数据专栏hive
1、评估磁盘空间大小、调整副本数、设置heapsize大小2、distcp-i-skipcrccheck源端到目标端,迁移3、元数据迁移,建表,替换location地址,或者导出db4、表分区修复5、配置增量T-1迁移或者T-26、校验历史分区脚本,表结构,大小,文件数7、根据ditcp不对的,进行补数脚本,删分区,重拉8、任务校验,客户跑完任务后,校验指定分区的count数和内容的md59、任务
- Redis Cluster集群详解
高冷小伙
redis数据库缓存
数据分片(Sharding)哈希槽(HashSlot)机制:集群将数据划分为16384个哈希槽,每个键通过CRC16(key)%16384计算归属的槽。槽分配给多个主节点,每个节点负责一部分槽(例如:3节点集群中,每个节点可能管理约5461个槽)。动态槽分配:节点增减时,槽可动态迁移,支持在线扩容/缩容(如CLUSTERADDSLOTS命令)。跨槽操作限制:事务、Lua脚本需确保所有键在同一槽(可
- 深入探讨Ceph:分布式存储架构的未来
深度Linux
ceph分布式架构C/C++
在数字化浪潮汹涌澎湃的当下,数据量呈爆发式增长,传统存储系统在应对海量数据存储、高并发访问以及灵活扩展等方面,逐渐显得力不从心。分布式存储技术应运而生,成为解决现代数据存储难题的关键方案,而Ceph作为分布式存储领域的佼佼者,正日益受到广泛关注和应用。Ceph以其卓越的性能、高可靠性、强大的扩展性以及开源的特性,在众多分布式存储系统中脱颖而出,被广泛应用于云计算、大数据、人工智能等前沿领域。无论是
- C++ STL std::vector 底层实现
zjkzjk7711
c++
C++STLstd::vector底层实现std::vector是C++STL中最常用的动态数组容器,其底层实现依赖于连续内存块,并采用动态扩容策略来管理内存。1.std::vector的底层数据结构std::vector内部维护三个指针templateclassvector{private:T*_start;//指向数据存储的起始位置T*_finish;//指向数据存储的末尾(size)T*_e
- 家政一城一店融合小程序怎么开通,需要哪些资质?
郑州拽牛科技
开源软件系统架构大数据小程序微信小程序
手把手教你开通洗衣洗鞋团购上门融合小程序!⚠️抖音新规重大调整!2025年起家政洗护必须"一城一店"(全国仅限365家连锁资质,地级市单店垄断!)开通秘籍三步走:✅核心资质:营业执照+法人身份证+商标注册证✅硬件证明:门头实拍图+室内全景视频+高德精准定位✅平台认证:ICP许可证+电信增值业务许可证(缺一不可!)遇到难题?90%商家都卡在这3个环节:1️⃣资质不全无法过审?2️⃣小程序功能不会搭建
- pytorch基础-layernormal 与 batchnormal
yuweififi
pytorch人工智能python
nn.LayerNorm(层归一化)和nn.BatchNorm(批量归一化)是深度学习中常用的两种归一化方法,都有助于提高模型的训练效率和稳定性,但它们在归一化维度、应用场景、计算方式等方面存在明显区别,以下为你详细介绍:1、归一化维度nn.LayerNorm:对单个样本的特征维度进行归一化。无论输入数据的形状如何,它会计算每个样本在特征维度上的均值和方差,然后进行归一化。例如,对于一个形状为(b
- anaconda 创建虚拟环境
yuweififi
环境搭建
1.打开AnacondaPrompt2.创建环境condacreate--nametorchpython=3.62.输入activatetorch安装的anacondapython虚拟环境打开,torch为创建的env名字3.condainfo--env查看所有创建的环境4.关闭环境deactivate切记先激活环境
- 通俗理解闭包
yuren_xia
前端技术javascript
JavaScript在ES6之前并没有类的概念,但通过原型链和闭包,开发者可以实现类似继承和封装的功能(原型链实现继承,闭包实现封装)。ES6引入了类语法,但闭包仍然是实现私有数据封装的重要手段之一。另外,使用闭包还可用于保存上下文信息等场景。一、定义从函数角度闭包是指有权访问另一个函数作用域中的变量的函数。即使外部函数已经返回,闭包仍然可以访问外部函数内部的变量。例如:functionouter
- 邻接矩阵存储图 C++题解
繁花开盛夏
图论算法开发语言c++图论
邻接矩阵存储图内存限制:128MiB时间限制:1000ms标准输入输出题目类型:传统评测方式:文本比较题目描述给出一个无向图,顶点数为n,边数为m。nusingnamespacestd;intn,m,x,y;inta[40][40];intmain(){scanf("%d%d",&n,&m);memset(a,0x3f,sizeof(a));for(inti=1;i<=m;i++){scanf("
- 【Multipath网络层协议】MPTCP工作原理
码上为赢
Multipath精通之路multipathMPTCP
常见网络层多路径协议介绍MPTCP(MultipathTCP)MPTCP是在传统TCP基础上进行扩展的协议,它允许在源端和目的端之间建立多个TCP子流,这些子流可以通过不同的网络路径传输数据。例如,一台笔记本电脑同时连接了Wi-Fi网络和以太网网络,当使用MPTCP进行数据传输时,它可以将数据分别通过Wi-Fi和以太网这两条不同的路径发送到目标服务器,从而充分利用两条链路的带宽。LISP(Loca
- 【面试实战】Spring基础、IoC、AOP、MVC、Mybatis、Spring Boot
Sivan_Xin
技术实战专栏(已上云)面试mvcspring
version:1.0文章目录SpringSpring基础/IoC♂️面试官:举例Spring的模块?♂️面试官:Spring、SpringMVC、SpringBoot关系?♂️面试官:说说对SpringIoC的了解?♂️面试官:什么是SpringBean?♂️面试官:Bean的作用域?♂️面试官:Bean的生命周期?♂️面试官:单例Bean的线程安全问题了解吗?♂️面试官:@Co
- 我在广州学 Mysql 系列——存储过程与存储函数详解
练小杰
数据库相关mysqlandroid数据库学习adbsql
ℹ️大家好,我是练小杰,今天周五了,一周就这样从手上溜走了,还有两星期过年!!本文将学习MYSQL中存储过程与存储函数的概念~~回顾:【索引详解】【索引相关练习】数据库专栏【数据库专栏】~想要了解更多内容,主页【练小杰的CSDN】文章目录存储过程与存储函数存储过程(StoredProcedure)存储函数(StoredFunction)⚠️主要区别选择存储过程还是存储函数创建存储过程命令解释创建存
- 我在广州学 Mysql 系列——数据表查询命令详解
练小杰
数据库相关mysql数据库学习经验分享adb后端
ℹ️大家好,我是LXJ,今天星期二了,本文将讲述MYSQL查询数据的详细命令以及相关例题~~复习:《Mysql函数的练习题》同时,数据库相关内容查看专栏【数据库专栏】~想要了解更多内容请点击我的主页:【练小杰的CSDN】“倒霉,倒霉,倒霉!”——龙叔文章目录前言基本查询语句单个表格查询查询所有字段查询指定字段查询指定记录带IN关键字的查询带BETWEENAND的范围查询带LIKE的字符匹配查询查询
- 通过TensorFlow实现简单深度学习模型(2)
yyc_audio
人工智能深度学习python机器学习
前文我们已经实现了对每批数据的训练,下面继续实现一轮完整的训练。完整的训练循环一轮训练就是对训练数据的每个批量都重复上述训练步骤,而完整的训练循环就是重复多轮训练。deffit(model,images,labels,epochs,batch_size=128):forepoch_counterinrange(epochs):print(f"Epoch{epoch_counter}")batch_
- c语言概率产生字母,智邮普创c语言面试题 ---- 字母概率(示例代码)
飞跃思考
c语言概率产生字母
题目描述小明最近对概率问题很感兴趣。一天,小明和小红一起玩一个概率游戏,首先小明给出一个字母和一个单词,然后由小红计算这个字母在这个单词中出现的概率。字母不区分大小写。例如,给定的字母是a,单词是apple,那么概率是0.20000。输入输入包含多组测试数据。每组数据包含一个字母和一个单词。单词的长度不超过200。输出对于每一个输入,输出对应的概率,结果保留5位小数。样例输入aapplecCand
- jQuery 跨域访问的三种方式 No 'Access-Control-Allow-Origin' header is present on the reque
qiaolevip
每天进步一点点学习永无止境跨域众观千象
XMLHttpRequest cannot load http://v.xxx.com. No 'Access-Control-Allow-Origin' header is present on the requested resource. Origin 'http://localhost:63342' is therefore not allowed access. test.html:1
- mysql 分区查询优化
annan211
java分区优化mysql
分区查询优化
引入分区可以给查询带来一定的优势,但同时也会引入一些bug.
分区最大的优点就是优化器可以根据分区函数来过滤掉一些分区,通过分区过滤可以让查询扫描更少的数据。
所以,对于访问分区表来说,很重要的一点是要在where 条件中带入分区,让优化器过滤掉无需访问的分区。
可以通过查看explain执行计划,是否携带 partitions
- MYSQL存储过程中使用游标
chicony
Mysql存储过程
DELIMITER $$
DROP PROCEDURE IF EXISTS getUserInfo $$
CREATE PROCEDURE getUserInfo(in date_day datetime)-- -- 实例-- 存储过程名为:getUserInfo-- 参数为:date_day日期格式:2008-03-08-- BEGINdecla
- mysql 和 sqlite 区别
Array_06
sqlite
转载:
http://www.cnblogs.com/ygm900/p/3460663.html
mysql 和 sqlite 区别
SQLITE是单机数据库。功能简约,小型化,追求最大磁盘效率
MYSQL是完善的服务器数据库。功能全面,综合化,追求最大并发效率
MYSQL、Sybase、Oracle等这些都是试用于服务器数据量大功能多需要安装,例如网站访问量比较大的。而sq
- pinyin4j使用
oloz
pinyin4j
首先需要pinyin4j的jar包支持;jar包已上传至附件内
方法一:把汉字转换为拼音;例如:编程转换后则为biancheng
/**
* 将汉字转换为全拼
* @param src 你的需要转换的汉字
* @param isUPPERCASE 是否转换为大写的拼音; true:转换为大写;fal
- 微博发送私信
随意而生
微博
在前面文章中说了如和获取登陆时候所需要的cookie,现在只要拿到最后登陆所需要的cookie,然后抓包分析一下微博私信发送界面
http://weibo.com/message/history?uid=****&name=****
可以发现其发送提交的Post请求和其中的数据,
让后用程序模拟发送POST请求中的数据,带着cookie发送到私信的接入口,就可以实现发私信的功能了。
- jsp
香水浓
jsp
JSP初始化
容器载入JSP文件后,它会在为请求提供任何服务前调用jspInit()方法。如果您需要执行自定义的JSP初始化任务,复写jspInit()方法就行了
JSP执行
这一阶段描述了JSP生命周期中一切与请求相关的交互行为,直到被销毁。
当JSP网页完成初始化后
- 在 Windows 上安装 SVN Subversion 服务端
AdyZhang
SVN
在 Windows 上安装 SVN Subversion 服务端2009-09-16高宏伟哈尔滨市道里区通达街291号
最佳阅读效果请访问原地址:http://blog.donews.com/dukejoe/archive/2009/09/16/1560917.aspx
现在的Subversion已经足够稳定,而且已经进入了它的黄金时段。我们看到大量的项目都在使
- android开发中如何使用 alertDialog从listView中删除数据?
aijuans
android
我现在使用listView展示了很多的配置信息,我现在想在点击其中一条的时候填出 alertDialog,点击确认后就删除该条数据,( ArrayAdapter ,ArrayList,listView 全部删除),我知道在 下面的onItemLongClick 方法中 参数 arg2 是选中的序号,但是我不知道如何继续处理下去 1 2 3
- jdk-6u26-linux-x64.bin 安装
baalwolf
linux
1.上传安装文件(jdk-6u26-linux-x64.bin)
2.修改权限
[root@localhost ~]# ls -l /usr/local/jdk-6u26-linux-x64.bin
3.执行安装文件
[root@localhost ~]# cd /usr/local
[root@localhost local]# ./jdk-6u26-linux-x64.bin&nbs
- MongoDB经典面试题集锦
BigBird2012
mongodb
1.什么是NoSQL数据库?NoSQL和RDBMS有什么区别?在哪些情况下使用和不使用NoSQL数据库?
NoSQL是非关系型数据库,NoSQL = Not Only SQL。
关系型数据库采用的结构化的数据,NoSQL采用的是键值对的方式存储数据。
在处理非结构化/半结构化的大数据时;在水平方向上进行扩展时;随时应对动态增加的数据项时可以优先考虑使用NoSQL数据库。
在考虑数据库的成熟
- JavaScript异步编程Promise模式的6个特性
bijian1013
JavaScriptPromise
Promise是一个非常有价值的构造器,能够帮助你避免使用镶套匿名方法,而使用更具有可读性的方式组装异步代码。这里我们将介绍6个最简单的特性。
在我们开始正式介绍之前,我们想看看Javascript Promise的样子:
var p = new Promise(function(r
- [Zookeeper学习笔记之八]Zookeeper源代码分析之Zookeeper.ZKWatchManager
bit1129
zookeeper
ClientWatchManager接口
//接口的唯一方法materialize用于确定那些Watcher需要被通知
//确定Watcher需要三方面的因素1.事件状态 2.事件类型 3.znode的path
public interface ClientWatchManager {
/**
* Return a set of watchers that should
- 【Scala十五】Scala核心九:隐式转换之二
bit1129
scala
隐式转换存在的必要性,
在Java Swing中,按钮点击事件的处理,转换为Scala的的写法如下:
val button = new JButton
button.addActionListener(
new ActionListener {
def actionPerformed(event: ActionEvent) {
- Android JSON数据的解析与封装小Demo
ronin47
转自:http://www.open-open.com/lib/view/open1420529336406.html
package com.example.jsondemo;
import org.json.JSONArray;
import org.json.JSONException;
import org.json.JSONObject;
impor
- [设计]字体创意设计方法谈
brotherlamp
UIui自学ui视频ui教程ui资料
从古至今,文字在我们的生活中是必不可少的事物,我们不能想象没有文字的世界将会是怎样。在平面设计中,UI设计师在文字上所花的心思和功夫最多,因为文字能直观地表达UI设计师所的意念。在文字上的创造设计,直接反映出平面作品的主题。
如设计一幅戴尔笔记本电脑的广告海报,假设海报上没有出现“戴尔”两个文字,即使放上所有戴尔笔记本电脑的图片都不能让人们得知这些电脑是什么品牌。只要写上“戴尔笔
- 单调队列-用一个长度为k的窗在整数数列上移动,求窗里面所包含的数的最大值
bylijinnan
java算法面试题
import java.util.LinkedList;
/*
单调队列 滑动窗口
单调队列是这样的一个队列:队列里面的元素是有序的,是递增或者递减
题目:给定一个长度为N的整数数列a(i),i=0,1,...,N-1和窗长度k.
要求:f(i) = max{a(i-k+1),a(i-k+2),..., a(i)},i = 0,1,...,N-1
问题的另一种描述就
- struts2处理一个form多个submit
chiangfai
struts2
web应用中,为完成不同工作,一个jsp的form标签可能有多个submit。如下代码:
<s:form action="submit" method="post" namespace="/my">
<s:textfield name="msg" label="叙述:">
- shell查找上个月,陷阱及野路子
chenchao051
shell
date -d "-1 month" +%F
以上这段代码,假如在2012/10/31执行,结果并不会出现你预计的9月份,而是会出现八月份,原因是10月份有31天,9月份30天,所以-1 month在10月份看来要减去31天,所以直接到了8月31日这天,这不靠谱。
野路子解决:假设当天日期大于15号
- mysql导出数据中文乱码问题
daizj
mysql中文乱码导数据
解决mysql导入导出数据乱码问题方法:
1、进入mysql,通过如下命令查看数据库编码方式:
mysql> show variables like 'character_set_%';
+--------------------------+----------------------------------------+
| Variable_name&nbs
- SAE部署Smarty出现:Uncaught exception 'SmartyException' with message 'unable to write
dcj3sjt126com
PHPsmartysae
对于SAE出现的问题:Uncaught exception 'SmartyException' with message 'unable to write file...。
官方给出了详细的FAQ:http://sae.sina.com.cn/?m=faqs&catId=11#show_213
解决方案为:
01
$path
- 《教父》系列台词
dcj3sjt126com
Your love is also your weak point.
你的所爱同时也是你的弱点。
If anything in this life is certain, if history has taught us anything, it is
that you can kill anyone.
不顾家的人永远不可能成为一个真正的男人。 &
- mongodb安装与使用
dyy_gusi
mongo
一.MongoDB安装和启动,widndows和linux基本相同
1.下载数据库,
linux:mongodb-linux-x86_64-ubuntu1404-3.0.3.tgz
2.解压文件,并且放置到合适的位置
tar -vxf mongodb-linux-x86_64-ubun
- Git排除目录
geeksun
git
在Git的版本控制中,可能有些文件是不需要加入控制的,那我们在提交代码时就需要忽略这些文件,下面讲讲应该怎么给Git配置一些忽略规则。
有三种方法可以忽略掉这些文件,这三种方法都能达到目的,只不过适用情景不一样。
1. 针对单一工程排除文件
这种方式会让这个工程的所有修改者在克隆代码的同时,也能克隆到过滤规则,而不用自己再写一份,这就能保证所有修改者应用的都是同一
- Ubuntu 创建开机自启动脚本的方法
hongtoushizi
ubuntu
转载自: http://rongjih.blog.163.com/blog/static/33574461201111504843245/
Ubuntu 创建开机自启动脚本的步骤如下:
1) 将你的启动脚本复制到 /etc/init.d目录下 以下假设你的脚本文件名为 test。
2) 设置脚本文件的权限 $ sudo chmod 755
- 第八章 流量复制/AB测试/协程
jinnianshilongnian
nginxluacoroutine
流量复制
在实际开发中经常涉及到项目的升级,而该升级不能简单的上线就完事了,需要验证该升级是否兼容老的上线,因此可能需要并行运行两个项目一段时间进行数据比对和校验,待没问题后再进行上线。这其实就需要进行流量复制,把流量复制到其他服务器上,一种方式是使用如tcpcopy引流;另外我们还可以使用nginx的HttpLuaModule模块中的ngx.location.capture_multi进行并发
- 电商系统商品表设计
lkl
DROP TABLE IF EXISTS `category`; -- 类目表
/*!40101 SET @saved_cs_client = @@character_set_client */;
/*!40101 SET character_set_client = utf8 */;
CREATE TABLE `category` (
`id` int(11) NOT NUL
- 修改phpMyAdmin导入SQL文件的大小限制
pda158
sqlmysql
用phpMyAdmin导入mysql数据库时,我的10M的
数据库不能导入,提示mysql数据库最大只能导入2M。
phpMyAdmin数据库导入出错: You probably tried to upload too large file. Please refer to documentation for ways to workaround this limit.
- Tomcat性能调优方案
Sobfist
apachejvmtomcat应用服务器
一、操作系统调优
对于操作系统优化来说,是尽可能的增大可使用的内存容量、提高CPU的频率,保证文件系统的读写速率等。经过压力测试验证,在并发连接很多的情况下,CPU的处理能力越强,系统运行速度越快。。
【适用场景】 任何项目。
二、Java虚拟机调优
应该选择SUN的JVM,在满足项目需要的前提下,尽量选用版本较高的JVM,一般来说高版本产品在速度和效率上比低版本会有改进。
J
- SQLServer学习笔记
vipbooks
数据结构xml
1、create database school 创建数据库school
2、drop database school 删除数据库school
3、use school 连接到school数据库,使其成为当前数据库
4、create table class(classID int primary key identity not null)
创建一个名为class的表,其有一