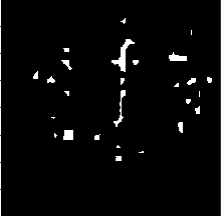医学图像分割--U-Net的颅内血管的分割 keras 实现
最近在进行医学图像的处理研究,主要是进行了颅内血管的CT图像的分割研究。
现阶段进行深度学习的图像分割的研究基本就是几种比较成熟的几个方法,FCN,u-net deeplab等一些方法。具体方法的操作,我这里就不细说了。大家可以自己去看看论文。
现阶段进行医学图像的深度学习处理的方法,最大的瓶颈就是数据量比较少,导致网络训练不是很理想。由于进行颅内血管的数据量也比较少,所以需要进行一定的数据量的扩充操作。由于颅内的CT图像是3D的数据。在这里,我为了使得进行更好的网络运行,我选择进行了切分操作,获得了2D数据。原来数据大小为242*512*512的,我进行了切割操作变成了400*400*200的数据量。
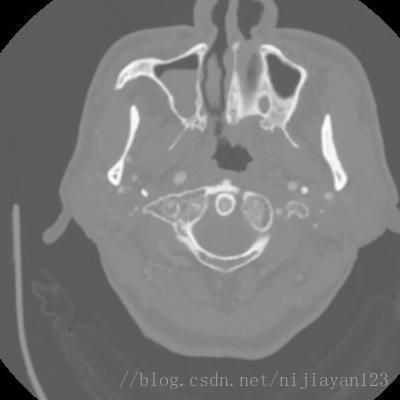
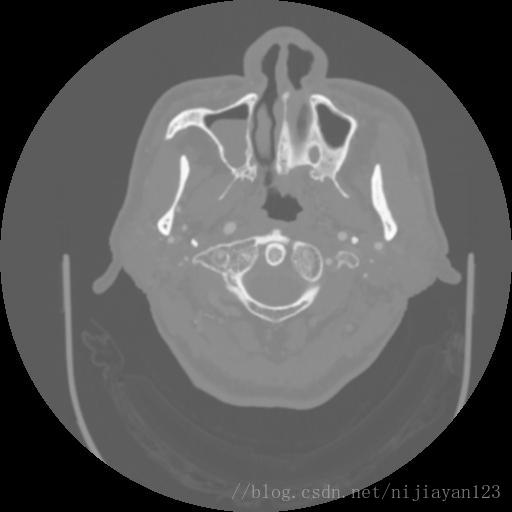
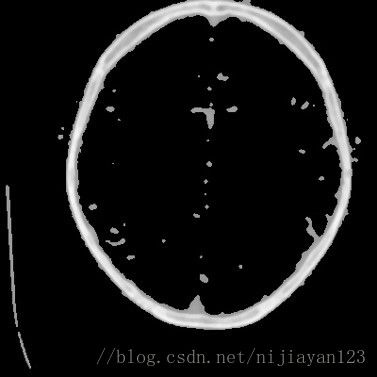
左边为剪切过的图片。
在进行剪切以后,由于进行医学图像中有大量不必要的信息量,所以在这里我还进行了二值化操作。


可以看到在进行二值化操作之后和ground true对比可以发现,大部分细节还是保持了。这样有利于进行后面进行卷积操作。
在这里我只是演示了一下整个进行操作的流程,如果还要进行精确的分割操作,还需要进行数据扩充操作,当然这些操作在keras中都有相关的操作。
在这里我使用的keras版本是2.1.1 tensorflow的版本是1.4.0。其他的版本是代码运行时什么情况我不知道。
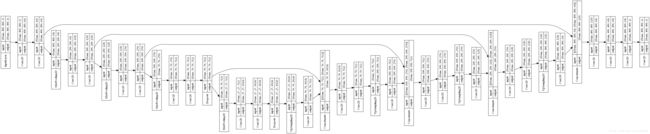
上述为U-net网络结构。
def dice_coef(y_true, y_pred):
smooth = 1.
y_true /= 255.
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
return (2. * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
def dice_coef_loss(y_true, y_pred):
return 1-dice_coef(y_true, y_pred)def unet():
inputs = Input((img_rows, img_cols,1))
conv1 = Conv2D(16, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(inputs)
conv1 = Conv2D(16, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv1)
pool1 = MaxPooling2D(pool_size=(2, 2))(conv1)
conv2 = Conv2D(32, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(pool1)
conv2 = Conv2D(32, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv2)
pool2 = MaxPooling2D(pool_size=(2, 2))(conv2)
conv3 = Conv2D(64, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(pool2)
conv3 = Conv2D(64, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv3)
pool3 = MaxPooling2D(pool_size=(2, 2))(conv3)
conv4 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(pool3)
conv4 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv4)
drop4 = Dropout(0.5)(conv4)
pool4 = MaxPooling2D(pool_size=(2, 2))(drop4)
conv5 = Conv2D(256, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(pool4)
conv5 = Conv2D(256, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv5)
drop5 = Dropout(0.5)(conv5)
upsame1=UpSampling2D(size = (2,2))(drop5)
up6 = Conv2D(128, 2, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(upsame1)
merge6 = concatenate([drop4,up6], axis = -1)
conv6 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(merge6)
conv6 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv6)
upsame2=UpSampling2D(size = (2,2))(conv6)
up7 = Conv2D(64, 2, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(upsame2)
merge7 = concatenate([conv3,up7],axis = -1)
conv7 = Conv2D(64, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(merge7)
conv7 = Conv2D(64, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv7)
up8 = Conv2D(128, 2, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(UpSampling2D(size = (2,2))(conv7))
merge8 = concatenate([conv2,up8], axis = -1)
conv8 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(merge8)
conv8 = Conv2D(128, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv8)
up9 = Conv2D(32, 2, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(UpSampling2D(size = (2,2))(conv8))
merge9 = concatenate([conv1,up9], axis = -1)
conv9 = Conv2D(32, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(merge9)
conv9 = Conv2D(32, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv9)
conv9 = Conv2D(2, 3, activation = 'relu', padding = 'same', kernel_initializer = 'he_normal')(conv9)
conv10 = Conv2D(1, 1, activation = 'sigmoid')(conv9)
model = Model(input = inputs, output = conv10)
model.summary()
model.compile(optimizer = Adam(lr = 0.0004), loss = dice_coef_loss, metrics = ['accuracy',dice_coef])
#plot_model(model, to_file='model1.png',show_shapes=True)
plot_model(model, to_file='model.png', show_shapes=True, show_layer_names=False)
return model在进行50次迭代以后准确率在94%左右。当然这只是一个简单的尝试,如果后续对图片进行扩充在进行训练可以达到的效果会好一点。在下图中可以发现整体的分割效果不是很理想。整个像素块比较大。后续还是要进行训练。
U-Net: Convolutional Networks for Biomedical Image Segmentation