Isolation Forest(二)之spark-iforest源码分析
github地址:https://github.com/titicaca/spark-iforest
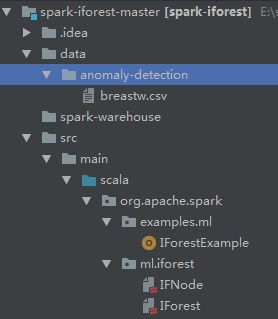
项目的目录结构如图,breastw.csv是乳腺癌分类数据
全部数据地址:https://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.data
数据![]()
1.示例代码编号ID编号
2.团块厚度1 - 10
3.细胞大小的均匀性1 - 10
4.细胞形状的均匀性1 - 10
5.边际粘合力1 - 10
6.单个上皮细胞大小1 - 10
7.裸核1 - 10
8.布兰德染色质1 - 10
9.普通核仁1 - 10
10.有丝分裂1 - 10
11.类:( 2为良性,4为恶性)
代码首先创建sparkSession,用于读取数据,初始化 new IForest(),然后开始fit训练数据
object IForestExample {
def main(args: Array[String]): Unit = {
val spark = SparkSession
.builder()
.master("local") // test in local mode
.appName("iforest example")
.getOrCreate()
val startTime = System.currentTimeMillis()
// Dataset from https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Original)
val dataset = spark.read.option("inferSchema", "true")
.csv("data/anomaly-detection/breastw.csv")
// Index label values: 2 -> 0, 4 -> 1
val indexer = new StringIndexer()
.setInputCol("_c10")
.setOutputCol("label")
val assembler = new VectorAssembler()
// assembler.setInputCols(Array("_c1", "_c2", "_c3", "_c4", "_c5", "_c6", "_c7", "_c8", "_c9"))
assembler.setInputCols(Array("_c1", "_c2", "_c3", "_c4", "_c5", "_c6", "_c7", "_c8", "_c9"))
assembler.setOutputCol("features")
val iForest = new IForest()
.setNumTrees(100)
.setMaxSamples(256)
.setContamination(0.35)
.setBootstrap(false)
.setMaxDepth(100)
.setSeed(123456L)
val pipeline = new Pipeline().setStages(Array(indexer, assembler, iForest))
val model = pipeline.fit(dataset)
val predictions = model.transform(dataset)
val binaryMetrics = new BinaryClassificationMetrics(
predictions.select("prediction", "label").rdd.map {
case Row(label: Double, ground: Double) => (label, ground)
}
)
val endTime = System.currentTimeMillis()
println(s"Training and predicting time: ${(endTime - startTime) / 1000} seconds.")
println(s"The model's auc: ${binaryMetrics.areaUnderROC()}")
}
}模型的训练
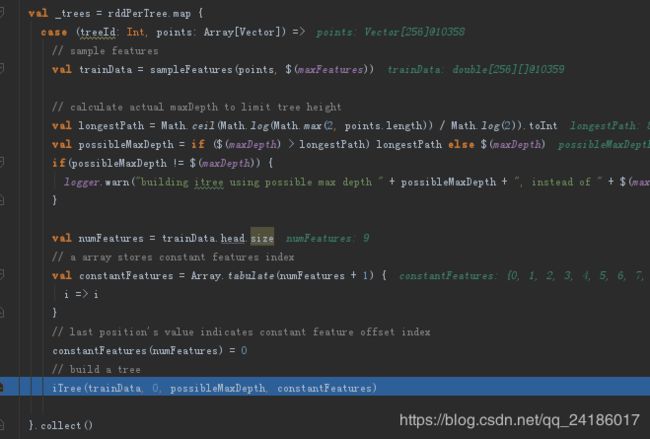
在pipeline.fit(dataset)最终调用iforest的fit方法,fit方法中构建森林,调用iTree函数创建树
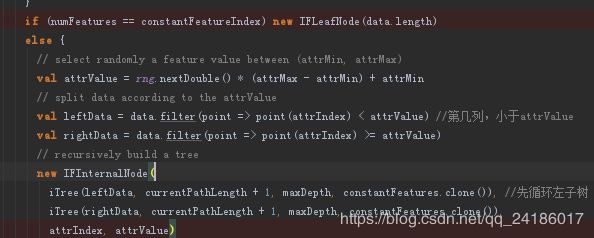
在iTree方法中,首先从训练特征属性中随机选取一个参数,再从features(attrIndex)选择这列的值,在这列的最大值和最小值之间选择一个随机数,所有数据中进行对比,将小于此随机数的放到一个dataset,将大于此随机数的放到一个dataset。然后返回内部节点,在内部节点中进行循环构成一棵二叉树。
然后进行预测,打标签
![]()
最后将模型保存
模型的预测
在val predictions = model.transform(dataset) 中进行预测,最后进行预测的代码在iForest中执行。
override def transform(dataset: Dataset[_]): DataFrame = {
transformSchema(dataset.schema, logging = true)
val numSamples = dataset.count() //列数
val possibleMaxSamples =
if ($(maxSamples) > 1.0) $(maxSamples) else ($(maxSamples) * numSamples)
val bcastModel = dataset.sparkSession.sparkContext.broadcast(this)
// calculate anomaly score
val scoreUDF = udf { (features: Vector) => { //一行
val normFactor = avgLength(possibleMaxSamples)
val avgPathLength = bcastModel.value.calAvgPathLength(features)
Math.pow(2, -avgPathLength / normFactor)
}
}
// append a score column
val scoreDataset = dataset.withColumn($(anomalyScoreCol), scoreUDF(col($(featuresCol))))
// get threshold value
val threshold = scoreDataset.stat.approxQuantile($(anomalyScoreCol),
Array(1 - $(contamination)), 0)
// set anomaly instance label 1
val predictUDF = udf { (anomalyScore: Double) =>
if (anomalyScore >= threshold(0)) 1.0 else 0.0
}
scoreDataset.withColumn($(predictionCol), predictUDF(col($(anomalyScoreCol))))
}
/**
* Calculate an average path length for a given feature set in a forest.
* @param features A Vector stores feature values.
* @return Average path length.
*/
private def calAvgPathLength(features: Vector): Double = {
val avgPathLength = trees.map(ifNode => { //100棵树,进行map,ifNode=iTree
calPathLength(features, ifNode, 0)
}).sum / trees.length
avgPathLength
}
/**
* Calculate a path langth for a given feature set in a tree.
* @param features A Vector stores feature values.
* @param ifNode Tree's root node.
* @param currentPathLength Current path length.
* @return Path length in this tree.
*/
private def calPathLength(features: Vector,
ifNode: IFNode,
currentPathLength: Int): Double = ifNode match {
case leafNode: IFLeafNode => currentPathLength + avgLength(leafNode.numInstance)
case internalNode: IFInternalNode =>
val attrIndex = internalNode.featureIndex
if (features(attrIndex) < internalNode.featureValue) {
calPathLength(features, internalNode.leftChild, currentPathLength + 1)
} else {
calPathLength(features, internalNode.rightChild, currentPathLength + 1)
}
}
/**
* A function to calculate an expected path length with a specific data size.
* @param size Data size.
* @return An expected path length.
*/
private def avgLength(size: Double): Double = {
if (size > 2) {
val H = Math.log(size - 1) + EulerConstant
2 * H - 2 * (size - 1) / size
}
else if (size == 2) 1.0
else 0.0
}