利用神经网络解决非线性回归问题
问题描述
现在我们通常使用神经网络进行分类,但是有时我们也会进行回归分析。
如本文的问题:
我们知道一个生物体内的原始有毒物质的量,然后对这个生物体进行治疗,向其体内注射一个物质,过一段时间后重新测量这个生物体内有毒物质量的多少。
因此,问题中有两个输入,都是标量数据,分别为有毒物质的量和注射物质的量,一个输出,也就是注射治疗物质后一段时间生物体的有毒物质的量。
数据如下图:
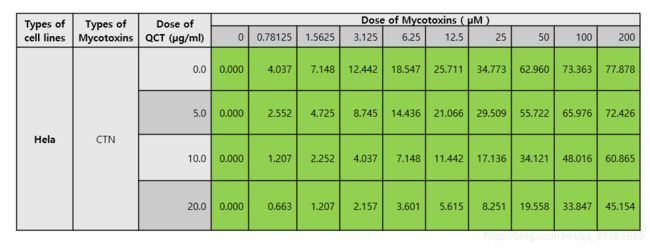
其中Dose of Mycotoxins 就是有毒物质,Dose of QCT就是治疗的药物。
其中蓝色底纹的数字就是输出结果。
一些说明
由于本文是进行回归分析,所以最后一层不进行激活,而直接输出。
本文程序使用sigmoid函数进行激活。
本文程序要求程序有一定的可重复性,隐含层可以指定。
另外,注意到
本文将使用数据预处理,也就是将数据减去均值再除以方差,否则使用sigmoid将会导致梯度消失。
因为数据比较大,比如200,这时输入200,当sigmoid函数的梯度就是接近于0了。
与此同时,我们在每一次激活前都进行BN处理,也就是batch normalize,中文可以翻译成规范化。
否则也会导致梯度消失的问题。与预处理情况相同。
程序
程序包括两部分,一部分是模型框架,一个是训练模型
第一部分:
# coding=utf-8
import numpy as np
def basic_forard(x, w, b):
x = x.reshape(x.shape[0], -1)
out = np.dot(x, w) + b
cache = (x, w, b)
return out, cache
def basic_backward(dout, cache):
x, w, b = cache
dout = np.array(dout)
dx = np.dot(dout, w.T)
# dx = np.reshape(dx, x.shape)
# x = x.reshape(x.shape[0], -1)
dw = np.dot(x.T, dout)
db = np.reshape(np.sum(dout, axis=0), b.shape)
return dx, dw, db
def batchnorm_forward(x, gamma, beta, bn_param):
mode = bn_param['mode']
eps = bn_param.get('eps', 1e-5)
momentum = bn_param.get('momentum', 0.9)
N, D = x.shape
running_mean = bn_param.get('running_mean', np.zeros(D, dtype=x.dtype))
running_var = bn_param.get('running_var', np.zeros(D, dtype=x.dtype))
out, cache = None, None
if mode == 'train':
sample_mean = np.mean(x, axis=0)
sample_var = np.var(x, axis=0)
x_hat = (x - sample_mean) / (np.sqrt(sample_var + eps))
out = gamma * x_hat + beta
cache = (gamma, x, sample_mean, sample_var, eps, x_hat)
running_mean = momentum * running_mean + (1 - momentum) * sample_mean
running_var = momentum * running_var + (1 - momentum) * sample_var
elif mode == 'test':
scale = gamma / (np.sqrt(running_var + eps))
out = x * scale + (beta - running_mean * scale)
else:
raise ValueError('Invalid forward batchnorm mode "%s"' % mode)
bn_param['running_mean'] = running_mean
bn_param['running_var'] = running_var
return out, cache
def batchnorm_backward(dout, cache):
gamma, x, u_b, sigma_squared_b, eps, x_hat = cache
N = x.shape[0]
dx_1 = gamma * dout
dx_2_b = np.sum((x - u_b) * dx_1, axis=0)
dx_2_a = ((sigma_squared_b + eps) ** -0.5) * dx_1
dx_3_b = (-0.5) * ((sigma_squared_b + eps) ** -1.5) * dx_2_b
dx_4_b = dx_3_b * 1
dx_5_b = np.ones_like(x) / N * dx_4_b
dx_6_b = 2 * (x - u_b) * dx_5_b
dx_7_a = dx_6_b * 1 + dx_2_a * 1
dx_7_b = dx_6_b * 1 + dx_2_a * 1
dx_8_b = -1 * np.sum(dx_7_b, axis=0)
dx_9_b = np.ones_like(x) / N * dx_8_b
dx_10 = dx_9_b + dx_7_a
dgamma = np.sum(x_hat * dout, axis=0)
dbeta = np.sum(dout, axis=0)
dx = dx_10
return dx, dgamma, dbeta
# def relu_forward(x):
# out = None
# out = np.maximum(0,x)
# cache = x
# return out, cache
#
#
# def relu_backward(dout, cache):
# dx, x = None, cache
# dx = (x >= 0) * dout
# return dx
def sigmoid_forward(x):
x = x.reshape(x.shape[0], -1)
out = 1 / (1 + np.exp(-1 * x))
cache = out
return out, cache
def sigmoid_backward(dout, cache):
out = cache
dx = out * (1 - out)
dx *= dout
return dx
def basic_sigmoid_forward(x, w, b):
basic_out, basic_cache = basic_forard(x, w, b)
sigmoid_out, sigmoid_cache = sigmoid_forward(basic_out)
cache = (basic_cache, sigmoid_cache)
return sigmoid_out, cache
# def basic_relu_forward(x, w, b):
# basic_out, basic_cache = basic_forard(x, w, b)
# relu_out, relu_cache = relu_forward(basic_out)
# cache = (basic_cache, relu_cache)
#
# return relu_out, cache
def basic_sigmoid_backward(dout, cache):
basic_cache, sigmoid_cache = cache
dx_sigmoid = sigmoid_backward(dout, sigmoid_cache)
dx, dw, db = basic_backward(dx_sigmoid, basic_cache)
return dx, dw, db
# def basic_relu_backward(dout, cache):
# basic_cache, relu_cache = cache
# dx_relu = relu_backward(dout, relu_cache)
# dx, dw, db = basic_backward(dx_relu, basic_cache)
#
# return dx, dw, db
def mean_square_error(x, y):
x = np.ravel(x)
loss = 0.5 * np.sum(np.square(y - x)) / x.shape[0]
dx = (x - y).reshape(-1, 1)
return loss, dx
class muliti_layer_net(object):
def __init__(self, hidden_dim, input_dim=2, num_classes=2, weight_scale=0.01, dtype=np.float32, seed=None, reg=0.0, use_batchnorm=True):
self.num_layers = 1 + len(hidden_dim)
self.dtype = dtype
self.reg = reg
self.params = {}
self.weight_scale = weight_scale
self.use_batchnorm = use_batchnorm
# init all parameters
layers_dims = [input_dim] + hidden_dim + [num_classes]
for i in range(self.num_layers):
self.params['W' + str(i + 1)] = np.random.randn(layers_dims[i], layers_dims[i + 1]) * self.weight_scale
self.params['b' + str(i + 1)] = np.zeros((1, layers_dims[i + 1]))
if self.use_batchnorm and i < (self.num_layers - 1):
self.params['gamma' + str(i + 1)] = np.ones((1, layers_dims[i + 1]))
self.params['beta' + str(i + 1)] = np.zeros((1, layers_dims[i + 1]))
self.bn_params = [] # list
if self.use_batchnorm:
self.bn_params = [{'mode': 'train'} for i in range(self.num_layers - 1)]
def loss(self, X, y=None):
X = X.astype(self.dtype)
mode = 'test' if y is None else 'train'
# compute the forward data and cache
basic_sigmoid_cache = {}
layer_out = {}
layer_out[0] = X
out_basic_forward, cache_basic_forward = {}, {}
out_bn, cache_bn = {}, {}
out_sigmoid_forward, cache_sigmoid_forward = {}, {}
for lay in range(self.num_layers - 1):
# print('lay: %f' % lay)
W = self.params['W' + str(lay + 1)]
b = self.params['b' + str(lay + 1)]
if self.use_batchnorm:
gamma, beta = self.params['gamma' + str(lay + 1)], self.params['beta' + str(lay + 1)]
out_basic_forward[lay], cache_basic_forward[lay] = basic_forard(np.array(layer_out[lay]), W, b)
out_bn[lay], cache_bn[lay] = batchnorm_forward(np.array(out_basic_forward[lay]), gamma, beta, self.bn_params[lay])
layer_out[lay + 1], cache_sigmoid_forward[lay] = sigmoid_forward(np.array(out_bn[lay]))
# = out_sigmoid_forward[lay]
else:
layer_out[lay+1], basic_sigmoid_cache[lay] = basic_sigmoid_forward(layer_out[lay], W, b)
score, basic_cache = basic_forard(layer_out[self.num_layers-1], self.params['W' + str(self.num_layers)], self.params['b' + str(self.num_layers)])
# print('Congratulations: Loss is computed successfully!')
if mode == 'test':
return score
# compute the gradient
grads = {}
loss, dscore = mean_square_error(score, y)
dx, dw, db = basic_backward(dscore, basic_cache)
grads['W' + str(self.num_layers)] = dw + self.reg * self.params['W' + str(self.num_layers)]
grads['b' + str(self.num_layers)] = db
loss += 0.5 * self.reg * np.sum(self.params['W' + str(self.num_layers)] * self.params['b' + str(self.num_layers)])
dbn, dsigmoid = {}, {}
for index in range(self.num_layers - 1):
lay = self.num_layers - 1 - index - 1
loss += 0.5 * self.reg * np.sum(self.params['W' + str(lay + 1)] * self.params['b' + str(lay + 1)])
if self.use_batchnorm:
dsigmoid[lay] = sigmoid_backward(dx, cache_sigmoid_forward[lay])
dbn[lay], grads['gamma' + str(lay + 1)], grads['beta' + str(lay + 1)] = batchnorm_backward(dsigmoid[lay], cache_bn[lay])
dx, grads['W' + str(lay + 1)], grads['b' + str(lay + 1)] = basic_backward(dbn[lay], cache_basic_forward[lay])
else:
dx, dw, db = basic_sigmoid_backward(dx, basic_sigmoid_cache[lay])
for lay in range(self.num_layers):
grads['W' + str(lay + 1)] += self.reg * self.params['W' + str(lay + 1)]
return loss, grads
def sgd_momentum(w, dw, config=None):
if config is None: config = {}
config.setdefault('learning_rate', 1e-2)
config.setdefault('momentum', 0.9)
v = config.get('velocity', np.zeros_like(w))
v = config['momentum'] * v - config['learning_rate'] * dw
next_w = w + v
config['velocity'] = v
return next_w, config
class Solver(object):
def __init__(self, model, data, **kwargs):
self.model = model
self.X_train = data['X_train']
self.y_train = data['y_train']
self.X_val = data['X_val']
self.y_val = data['y_val']
self.update_rule = kwargs.pop('update_rule', 'sgd_momentum')
self.optim_config = kwargs.pop('optim_config', {})
self.lr_decay = kwargs.pop('lr_decay', 1.0)
self.batch_size = kwargs.pop('batch_size', 100)
self.num_epochs = kwargs.pop('num_epochs', 10)
self.weight_scale = kwargs.pop('weight_scale', 0.01)
self.print_every = kwargs.pop('print_every', 10)
self.verbose = kwargs.pop('verbose', True)
if len(kwargs) > 0:
extra = ', '.join('"%s"' % k for k in kwargs.keys())
raise ValueError('Unrecognized argements %s' % extra)
self._reset()
def _reset(self):
self.epoch = 100
self.best_val_acc = 0
self.best_params = {}
self.loss_history = []
self.train_acc_history = []
self.val_acc_history = []
self.optim_configs = {}
for p in self.model.params:
d = {k: v for k, v in self.optim_config.items()}
self.optim_configs[p] = d
def _step(self):
loss, grads = self.model.loss(self.X_train, self.y_train)
self.loss_history.append(loss)
for p, w in self.model.params.items():
dw = grads[p]
config = self.optim_configs[p]
next_w, next_config = sgd_momentum(w, dw, config)
self.model.params[p] = next_w
self.optim_configs[p] = next_config
return loss
def train(self):
min_loss = 100000000
num_train = self.X_train.shape[0]
iterations_per_epoch = max(num_train / self.batch_size, 1)
num_iterations = self.num_epochs * iterations_per_epoch
for t in range(int(num_iterations)):
loss = self._step()
if self.verbose:
# print(self.loss_history[-1])
pass
if loss < min_loss:
min_loss = loss
for k, v in self.model.params.items():
self.best_params[k] = v.copy()
self.model.params = self.best_params
第二部分
import numpy as np
# import data
dose_QCT = np.array([0, 5, 10, 20])
mean_QCT, std_QCT = np.mean(dose_QCT), np.std(dose_QCT)
dose_QCT = (dose_QCT - mean_QCT ) / std_QCT
dose_toxins = np.array([0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, 200])
mean_toxins, std_toxins = np.mean(dose_toxins), np.std(dose_toxins)
dose_toxins = (dose_toxins - mean_toxins ) / std_toxins
result = np.array([[0, 4.037, 7.148, 12.442, 18.547, 25.711, 34.773, 62.960, 73.363, 77.878],
[0, 2.552, 4.725, 8.745, 14.436, 21.066, 29.509, 55.722, 65.976, 72.426],
[0, 1.207, 2.252, 4.037, 7.148, 11.442, 17.136, 34.121, 48.016, 60.865],
[0, 0.663, 1.207, 2.157, 3.601, 5.615, 8.251, 19.558, 33.847, 45.154]])
mean_result, std_result = np.mean(result), np.std(result)
result = (result - mean_result ) / std_result
# create the train data
train_x, train_y = [], []
for i,qct in enumerate(dose_QCT):
for j,toxin in enumerate(dose_toxins):
x = [qct, toxin]
y = result[i, j]
train_x.append(x)
train_y.append(y)
train_x = np.array(train_x)
train_y = np.array(train_y)
print(train_x.shape)
print(train_y.shape)
import layers_regression
small_data = {'X_train': train_x,
'y_train': train_y,
'X_val': train_x,
'y_val': train_y,}
batch_size = train_x.shape[0]
learning_rate = 0.002
reg = 0
model = layers_regression.muliti_layer_net(hidden_dim=[5,5], input_dim=2, num_classes=1, reg=reg, dtype=np.float64)
solver = layers_regression.Solver(model, small_data, print_every=0, num_epochs=50000, batch_size=batch_size, weight_scale=1,
update_rule='sgd_momentum', optim_config={'learning_rate': learning_rate})
print('Please wait several minutes!')
solver.train()
# print(model.params)
best_model = model
print('Train process is finised')
import matplotlib.pyplot as plt
# %matplotlib inline
plt.plot(solver.loss_history, '.')
plt.title('Training loss history')
plt.xlabel('Iteration')
plt.ylabel('Training loss')
plt.show()
# predict the training_data
predict = best_model.loss(train_x)
predict = np.round(predict * std_result + mean_result, 1)
print('Predict is ')
print('{}'.format(predict.reshape(4, 10)))
# print('{}'.format(predict))
# observe the error between the predict after training with ground truth
result = np.array([[0, 4.037, 7.148, 12.442, 18.547, 25.711, 34.773, 62.960, 73.363, 77.878],
[0, 2.552, 4.725, 8.745, 14.436, 21.066, 29.509, 55.722, 65.976, 72.426],
[0, 1.207, 2.252, 4.037, 7.148, 11.442, 17.136, 34.121, 48.016, 60.865],
[0, 0.663, 1.207, 2.157, 3.601, 5.615, 8.251, 19.558, 33.847, 45.154]])
result = result.reshape(4, 10)
predict = predict.reshape(4, 10)
error = np.round(result - predict, 2)
print('error between predict and real data')
print(error)
print('The absulate error in all data is %f' % np.sum(np.abs(error)))
print('The mean error in all data is %f' % np.mean(np.abs(error)))
# figure the predict map in 3D
x_1 = (np.arange(0, 20, 0.1) - mean_QCT) / std_QCT
x_2 = (np.arange(0, 200, 1) - mean_toxins) / std_toxins
x_test = np.zeros((len(x_1)*len(x_2), 2))
index = 0
for i in range(len(x_1)):
for j in range(len(x_2)):
x_test[int(index), 0] = x_1[int(i)]
x_test[int(index), 1] = x_2[int(j)]
index += 1
test_pred = best_model.loss(x_test)
predict = np.round(test_pred * std_result + mean_result, 3)
from mpl_toolkits.mplot3d import Axes3D
x_1, x_2 = np.meshgrid(x_1 * std_QCT + mean_QCT, x_2 * std_toxins + mean_toxins)
figure = plt.figure()
ax = Axes3D(figure)
predict = predict.reshape(len(x_1), len(x_2))
ax.plot_surface(x_1, x_2, predict, rstride=1, cstride=1, cmap='rainbow')
plt.show()
# 最后本文将进行一些预测,但预测效果不是很好
# question 2: predict with given
dose_QCT_predict = np.ravel(np.array([7.5, 15]))
dose_QCT_predict_ = (dose_QCT_predict - mean_QCT)/ std_QCT
dose_toxins_predict = np.array([0, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, 200])
dose_toxins_predict_ = (dose_toxins_predict - mean_toxins) / std_toxins
test = []
for i,qct in enumerate(dose_QCT_predict):
for j,toxin in enumerate(dose_toxins_predict):
x = [qct, toxin]
test.append(x)
test = np.array(test)
print('Please look at the test data:')
print(test)
test = []
for i,qct in enumerate(dose_QCT_predict_):
for j,toxin in enumerate(dose_toxins_predict_):
x = [qct, toxin]
test.append(x)
test = np.array(test)
test_pred = best_model.loss(test)
predict = np.round(test_pred * std_result + mean_result, 1)
print(predict.reshape(2, 10))