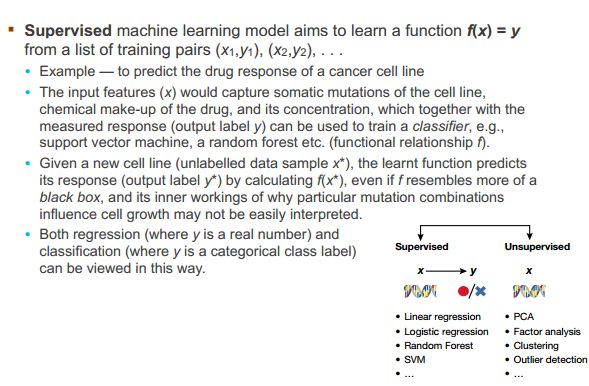
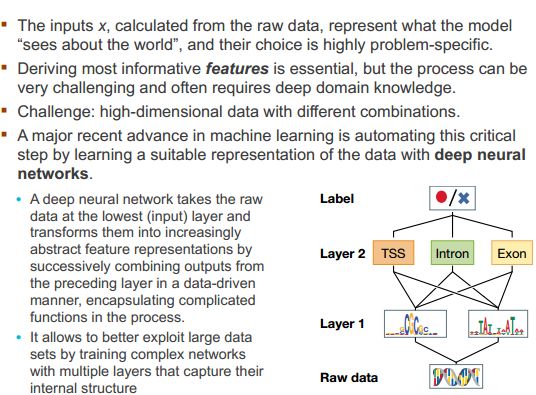
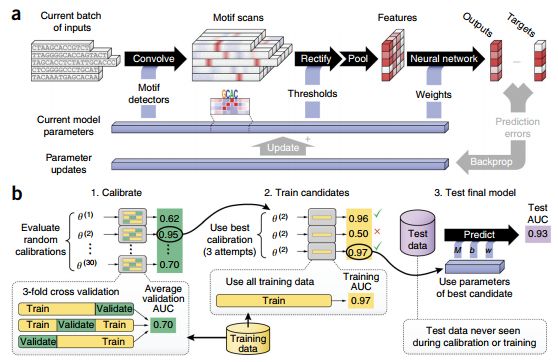
一、什么是宏函数?通过宏定义的函数是宏函数。如下,编译器在预处理阶段会将Add(x,y)替换为((x)*(y))#defineAdd(x,y)((x)*(y))#defineAdd(x,y)((x)*(y))intmain(){inta=10;intb=20;intd=10;intc=Add(a+d,b)*2;cout<
C语言如何定义宏函数?
小九格物
c语言
在C语言中,宏函数是通过预处理器定义的,它在编译之前替换代码中的宏调用。宏函数可以模拟函数的行为,但它们不是真正的函数,因为它们在编译时不会进行类型检查,也不会分配存储空间。宏函数的定义通常使用#define指令,后面跟着宏的名称和参数列表,以及宏展开后的代码。宏函数的定义方式:1.基本宏函数:这是最简单的宏函数形式,它直接定义一个表达式。#defineSQUARE(x)((x)*(x))2.带参
微服务下功能权限与数据权限的设计与实现
nbsaas-boot
微服务java架构
在微服务架构下,系统的功能权限和数据权限控制显得尤为重要。随着系统规模的扩大和微服务数量的增加,如何保证不同用户和服务之间的访问权限准确、细粒度地控制,成为设计安全策略的关键。本文将讨论如何在微服务体系中设计和实现功能权限与数据权限控制。1.功能权限与数据权限的定义功能权限:指用户或系统角色对特定功能的访问权限。通常是某个用户角色能否执行某个操作,比如查看订单、创建订单、修改用户资料等。数据权限:
理解Gunicorn:Python WSGI服务器的基石
范范0825
ipythonlinux运维
理解Gunicorn:PythonWSGI服务器的基石介绍Gunicorn,全称GreenUnicorn,是一个为PythonWSGI(WebServerGatewayInterface)应用设计的高效、轻量级HTTP服务器。作为PythonWeb应用部署的常用工具,Gunicorn以其高性能和易用性著称。本文将介绍Gunicorn的基本概念、安装和配置,帮助初学者快速上手。1.什么是Gunico
小丽成长记(四十三)
玲玲54321
小丽发现,即使她好不容易调整好自己的心态下一秒总会有不确定的伤脑筋的事出现,一个接一个的问题,人生就没有停下的时候,小问题不断出现。不过她今天看的书,她接受了人生就是不确定的,厉害的人就是不断创造确定性,在Ta的领域比别人多的确定性就能让自己脱颖而出,显示价值从而获得的比别人多的利益。正是这样的原因,因为从前修炼自己太少,使得她现在在人生道路上打怪起来困难重重,她似乎永远摆脱不了那种无力感,有种习
2021年12月19日,春蕾教育集团团建活动感受——黄晓丹
黄错错加油
感受:1.从陌生到熟悉的过程。游戏环节让我们在轻松的氛围中得到了锻炼,也增长了不少知识。2.游戏过程中,我们贡献的是个人力量,展现的是团队的力量。它磨合的往往不止是工作的熟悉,更是观念上契合度的贴近。3.这和工作是一样的道理。在各自的岗位上,每个人摆正自己的位置、各司其职充分发挥才能,并团结一致劲往一处使,才能实现最大的成功。新知:1.团队精神需要不断地创新。过去,人们把创新看作是冒风险,现在人们
c++ 的iostream 和 c++的stdio的区别和联系
黄卷青灯77
c++算法开发语言iostreamstdio
在C++中,iostream和C语言的stdio.h都是用于处理输入输出的库,但它们在设计、用法和功能上有许多不同。以下是两者的区别和联系:区别1.编程风格iostream(C++风格):C++标准库中的输入输出流类库,支持面向对象的输入输出操作。典型用法是cin(输入)和cout(输出),使用>操作符来处理数据。更加类型安全,支持用户自定义类型的输入输出。#includeintmain(){in
爬山后遗症
璃绛
爬山,攀登,一步一步走向制高点,是一种挑战。成功抵达是一种无法言语的快乐,在山顶吹吹风,看看风景,这是从未有过的体验。然而,爬山一时爽,下山腿打颤,颠簸的路,一路向下走,腿部力量不够,走起来抖到不行,停不下来了!第二天必定腿疼,浑身酸痛,坐立难安!
Long类型前后端数据不一致
igotyback
前端
响应给前端的数据浏览器控制台中response中看到的Long类型的数据是正常的到前端数据不一致前后端数据类型不匹配是一个常见问题,尤其是当后端使用Java的Long类型(64位)与前端JavaScript的Number类型(最大安全整数为2^53-1,即16位)进行数据交互时,很容易出现精度丢失的问题。这是因为JavaScript中的Number类型无法安全地表示超过16位的整数。为了解决这个问
swagger访问路径
igotyback
swagger
Swagger2.x版本访问地址:http://{ip}:{port}/{context-path}/swagger-ui.html{ip}是你的服务器IP地址。{port}是你的应用服务端口,通常为8080。{context-path}是你的应用上下文路径,如果应用部署在根路径下,则为空。Swagger3.x版本对于Swagger3.x版本(也称为OpenAPI3)访问地址:http://{ip
mysql禁用远程登录
igotyback
mysql
去mysql库中的user表里,将host都改成localhost之后刷新权限FLUSHPRIVILEGES;
如何在 Fork 的 GitHub 项目中保留自己的修改并同步上游更新?github_fork_update
iBaoxing
github
如何在Fork的GitHub项目中保留自己的修改并同步上游更新?在GitHub上Fork了一个项目后,你可能会对项目进行一些修改,同时原作者也在不断更新。如果想要在保留自己修改的基础上,同步原作者的最新更新,很多人会不知所措。本文将详细讲解如何在不丢失自己改动的情况下,将上游仓库的更新合并到自己的仓库中。问题描述假设你在GitHub上Fork了一个项目,并基于该项目做了一些修改,随后你发现原作者对
Linux下QT开发的动态库界面弹出操作(SDL2)
13jjyao
QT类qt开发语言sdl2linux
需求:操作系统为linux,开发框架为qt,做成需带界面的qt动态库,调用方为java等非qt程序难点:调用方为java等非qt程序,也就是说调用方肯定不带QApplication::exec(),缺少了这个,QTimer等事件和QT创建的窗口将不能弹出(包括opencv也是不能弹出);这与qt调用本身qt库是有本质的区别的思路:1.调用方缺QApplication::exec(),那么我们在接口
30天风格练习-DAY2
黄希夷
Day2(重义)在一个周日/一周的最后一天,我来到位于市中心/市区繁华地带的一家购物中心/商场,中心内人很多/熙熙攘攘。我注意到/看见一个独行/孤身一人的年轻女孩/,留着一头引人注目/长过腰际的头发,上身穿一件暗红色/比正红色更深的衣服/穿在身体上的东西。走下扶梯的时候,她摔倒了/跌向地面,在她正要站起来/让身体离开地面的时候,过长/超过一般人长度的头发被支撑身体/躯干的手掌压/按在下面,她赶紧用
店群合一模式下的社区团购新发展——结合链动 2+1 模式、AI 智能名片与 S2B2C 商城小程序源码
说私域
人工智能小程序
摘要:本文探讨了店群合一的社区团购平台在当今商业环境中的重要性和优势。通过分析店群合一模式如何将互联网社群与线下终端紧密结合,阐述了链动2+1模式、AI智能名片和S2B2C商城小程序源码在这一模式中的应用价值。这些创新元素的结合为社区团购带来了新的机遇,提升了用户信任感、拓展了营销渠道,并实现了线上线下的完美融合。一、引言随着互联网技术的不断发展,社区团购作为一种新兴的商业模式,在满足消费者日常需
我校举行新老教师师徒结对仪式暨名师专业工作室工作交流活动
李蕾1229
为促进我校教师专业发展,发挥骨干教师的引领带头作用,11月6日下午,我校举行新老教师师徒结对仪式暨名师专业工作室工作交流活动。图片发自App会议由教师发展处李蕾主任主持,首先,由范校长宣读新老教师结对名单及双方承担职责。随后,两位新调入教师陈玉萍、莫正杰分别和他们的师傅鲍元美、刘召彬老师签订了师徒结对协议书。图片发自App图片发自App师徒拥抱、握手。有了师傅就有了目标有了方向,相信两位新教师在师
向内而求
陈陈_19b4
10月27日,阴。阅读书目:《次第花开》。作者:希阿荣博堪布,是当今藏传佛家宁玛派最伟大的上师法王,如意宝晋美彭措仁波切颇具影响力的弟子之一。多年以来,赴海内外各地弘扬佛法,以正式授课、现场开示、发表文章等多种方法指导佛学弟子修行佛法。代表作《寂静之道》、《生命这出戏》、《透过佛法看世界》自出版以来一直是佛教类书籍中的畅销书。图片发自App金句:1.佛陀说,一切痛苦的根源在于我们长期以来对自身及外
消息中间件有哪些常见类型
xmh-sxh-1314
java
消息中间件根据其设计理念和用途,可以大致分为以下几种常见类型:点对点消息队列(Point-to-PointMessagingQueues):在这种模型中,消息被发送到特定的队列中,消费者从队列中取出并处理消息。队列中的消息只能被一个消费者消费,消费后即被删除。常见的实现包括IBM的MQSeries、RabbitMQ的部分使用场景等。适用于任务分发、负载均衡等场景。发布/订阅消息模型(Pub/Sub
ArcGIS栅格计算器常见公式(赋值、0和空值的转换、补充栅格空值)
研学随笔
arcgis经验分享
我们在使用ArcGIS时通常经常用到栅格计算器,今天主要给大家介绍我日常中经常用到的几个公式,供大家参考学习。将特定值(-9999)赋值为0,例如-9999.Con("raster"==-9999,0,"raster")2.给空值赋予特定的值(如0)Con(IsNull("raster"),0,"raster")3.将特定的栅格值(如1)赋值为空值,其他保留原值SetNull("raster"==
那个抄袭的大张伟
猫小努
最近一直在追《即刻电音》这个综艺,除了觉得出场节目的音乐制作人有意思之外,也觉得有两个导师挺有趣的(另外一个就忽略了吧)。孙艺兴在上一篇文章里面已经说过了,那么这篇就说说我们的大老师,大张伟吧。其实在节目刚开始大张伟出来的时候,我以为他是属于导师里面来活跃气氛负责搞笑的,毕竟孙艺兴属于卖萌卖傻卖老实的,尚雯婕一般负责装逼耍狠的,而大张伟一贯以来上综艺的形象基本上都是蹦蹦跳跳带动气氛的。谁知道,两期
三大师传
beca酱
巴尔扎克的作品被誉为“法国社会的一面镜子”。文学大师维克多·雨果对巴尔扎克的评价是:“在最伟大的人物中间,巴尔扎克是名列前茅者;在最优秀的人物中间,巴尔扎克是佼佼者之一。”一个原本寂寂无名的小人物,从地中海的某个海岛上,只身一人来到巴黎,没有朋友,也没有名望。作为一个一文不名的外乡人,凭着赤手空拳赢得了巴黎,征服了整个法兰西,并且赢得了世界。这个人就是十九世纪法国伟大的军事家、政治家,法兰西第一帝
2018-07-23-催眠日作业-#不一样的31天#-66小鹿
小鹿_33
预言日:人总是在逃避命运的路上,与之不期而遇。心理学上有个著名的名词,叫做自证预言;经济学上也有一个很著名的定律叫做,墨菲定律;在灵修派上,还有一个很著名的法则,叫做吸引力法则。这3个领域的词,虽然看起来不太一样,但是他们都在告诉人们一个现象:你越担心什么,就越有可能会发生什么。同样的道理,你越想得到什么,就应该要积极地去创造什么。无论是自证预言,墨菲定律还是吸引力法则,对人都有正反2个维度的影响
ios内付费
374016526
ios内付费
近年来写了很多IOS的程序,内付费也用到不少,使用IOS的内付费实现起来比较麻烦,这里我写了一个简单的内付费包,希望对大家有帮助。
具体使用如下:
这里的sender其实就是调用者,这里主要是为了回调使用。
[KuroStoreApi kuroStoreProductId:@"产品ID" storeSender:self storeFinishCallBa
20 款优秀的 Linux 终端仿真器
brotherlamp
linuxlinux视频linux资料linux自学linux教程
终端仿真器是一款用其它显示架构重现可视终端的计算机程序。换句话说就是终端仿真器能使哑终端看似像一台连接上了服务器的客户机。终端仿真器允许最终用户用文本用户界面和命令行来访问控制台和应用程序。(LCTT 译注:终端仿真器原意指对大型机-哑终端方式的模拟,不过在当今的 Linux 环境中,常指通过远程或本地方式连接的伪终端,俗称“终端”。)
你能从开源世界中找到大量的终端仿真器,它们
Solr Deep Paging(solr 深分页)
eksliang
solr深分页solr分页性能问题
转载请出自出处:http://eksliang.iteye.com/blog/2148370
作者:eksliang(ickes) blg:http://eksliang.iteye.com/ 概述
长期以来,我们一直有一个深分页问题。如果直接跳到很靠后的页数,查询速度会比较慢。这是因为Solr的需要为查询从开始遍历所有数据。直到Solr的4.7这个问题一直没有一个很好的解决方案。直到solr
数据库面试题
18289753290
面试题 数据库
1.union ,union all
网络搜索出的最佳答案:
union和union all的区别是,union会自动压缩多个结果集合中的重复结果,而union all则将所有的结果全部显示出来,不管是不是重复。
Union:对两个结果集进行并集操作,不包括重复行,同时进行默认规则的排序;
Union All:对两个结果集进行并集操作,包括重复行,不进行排序;
2.索引有哪些分类?作用是
Android TV屏幕适配
酷的飞上天空
android
先说下现在市面上TV分辨率的大概情况
两种分辨率为主
1.720标清,分辨率为1280x720.
屏幕尺寸以32寸为主,部分电视为42寸
2.1080p全高清,分辨率为1920x1080
屏幕尺寸以42寸为主,此分辨率电视屏幕从32寸到50寸都有
适配遇到问题,已1080p尺寸为例:
分辨率固定不变,屏幕尺寸变化较大。
如:效果图尺寸为1920x1080,如果使用d
Timer定时器与ActionListener联合应用
永夜-极光
java
功能:在控制台每秒输出一次
代码:
package Main;
import javax.swing.Timer;
import java.awt.event.*;
public class T {
private static int count = 0;
public static void main(String[] args){
Ubuntu14.04系统Tab键不能自动补全问题解决
随便小屋
Ubuntu 14.04
Unbuntu 14.4安装之后就在终端中使用Tab键不能自动补全,解决办法如下:
1、利用vi编辑器打开/etc/bash.bashrc文件(需要root权限)
sudo vi /etc/bash.bashrc
接下来会提示输入密码
2、找到文件中的下列代码
#enable bash completion in interactive shells
#if
学会人际关系三招 轻松走职场
aijuans
职场
要想成功,仅有专业能力是不够的,处理好与老板、同事及下属的人际关系也是门大学问。如何才能在职场如鱼得水、游刃有余呢?在此,教您简单实用的三个窍门。
第一,多汇报
最近,管理学又提出了一个新名词“追随力”。它告诉我们,做下属最关键的就是要多请示汇报,让上司随时了解你的工作进度,有了新想法也要及时建议。不知不觉,你就有了“追随力”,上司会越来越了解和信任你。
第二,勤沟通
团队的力
《O2O:移动互联网时代的商业革命》读书笔记
aoyouzi
读书笔记
移动互联网的未来:碎片化内容+碎片化渠道=各式精准、互动的新型社会化营销。
O2O:Online to OffLine 线上线下活动
O2O就是在移动互联网时代,生活消费领域通过线上和线下互动的一种新型商业模式。
手机二维码本质:O2O商务行为从线下现实世界到线上虚拟世界的入口。
线上虚拟世界创造的本意是打破信息鸿沟,让不同地域、不同需求的人
js实现图片随鼠标滚动的效果
百合不是茶
JavaScript滚动属性的获取图片滚动属性获取页面加载
1,获取样式属性值
top 与顶部的距离
left 与左边的距离
right 与右边的距离
bottom 与下边的距离
zIndex 层叠层次
例子:获取左边的宽度,当css写在body标签中时
<div id="adver" style="position:absolute;top:50px;left:1000p
ajax同步异步参数async
bijian1013
jqueryAjaxasync
开发项目开发过程中,需要将ajax的返回值赋到全局变量中,然后在该页面其他地方引用,因为ajax异步的原因一直无法成功,需将async:false,使其变成同步的。
格式:
$.ajax({ type: 'POST', ur
Webx3框架(1)
Bill_chen
eclipsespringmaven框架ibatis
Webx是淘宝开发的一套Web开发框架,Webx3是其第三个升级版本;采用Eclipse的开发环境,现在支持java开发;
采用turbine原型的MVC框架,扩展了Spring容器,利用Maven进行项目的构建管理,灵活的ibatis持久层支持,总的来说,还是一套很不错的Web框架。
Webx3遵循turbine风格,velocity的模板被分为layout/screen/control三部
【MongoDB学习笔记五】MongoDB概述
bit1129
mongodb
MongoDB是面向文档的NoSQL数据库,尽量业界还对MongoDB存在一些质疑的声音,比如性能尤其是查询性能、数据一致性的支持没有想象的那么好,但是MongoDB用户群确实已经够多。MongoDB的亮点不在于它的性能,而是它处理非结构化数据的能力以及内置对分布式的支持(复制、分片达到的高可用、高可伸缩),同时它提供的近似于SQL的查询能力,也是在做NoSQL技术选型时,考虑的一个重要因素。Mo
spring/hibernate/struts2常见异常总结
白糖_
Hibernate
Spring
①ClassNotFoundException: org.aspectj.weaver.reflect.ReflectionWorld$ReflectionWorldException
缺少aspectjweaver.jar,该jar包常用于spring aop中
②java.lang.ClassNotFoundException: org.sprin
jquery easyui表单重置(reset)扩展思路
bozch
formjquery easyuireset
在jquery easyui表单中 尚未提供表单重置的功能,这就需要自己对其进行扩展。
扩展的时候要考虑的控件有:
combo,combobox,combogrid,combotree,datebox,datetimebox
需要对其添加reset方法,reset方法就是把初始化的值赋值给当前的组件,这就需要在组件的初始化时将值保存下来。
在所有的reset方法添加完毕之后,就需要对fo
编程之美-烙饼排序
bylijinnan
编程之美
package beautyOfCoding;
import java.util.Arrays;
/*
*《编程之美》的思路是:搜索+剪枝。有点像是写下棋程序:当前情况下,把所有可能的下一步都做一遍;在这每一遍操作里面,计算出如果按这一步走的话,能不能赢(得出最优结果)。
*《编程之美》上代码有很多错误,且每个变量的含义令人费解。因此我按我的理解写了以下代码:
*/
Struts1.X 源码分析之ActionForm赋值原理
chenbowen00
struts
struts1在处理请求参数之前,首先会根据配置文件action节点的name属性创建对应的ActionForm。如果配置了name属性,却找不到对应的ActionForm类也不会报错,只是不会处理本次请求的请求参数。
如果找到了对应的ActionForm类,则先判断是否已经存在ActionForm的实例,如果不存在则创建实例,并将其存放在对应的作用域中。作用域由配置文件action节点的s
[空天防御与经济]在获得充足的外部资源之前,太空投资需有限度
comsci
资源
这里有一个常识性的问题:
地球的资源,人类的资金是有限的,而太空是无限的.....
就算全人类联合起来,要在太空中修建大型空间站,也不一定能够成功,因为资源和资金,技术有客观的限制....
&
ORACLE临时表—ON COMMIT PRESERVE ROWS
daizj
oracle临时表
ORACLE临时表 转
临时表:像普通表一样,有结构,但是对数据的管理上不一样,临时表存储事务或会话的中间结果集,临时表中保存的数据只对当前
会话可见,所有会话都看不到其他会话的数据,即使其他会话提交了,也看不到。临时表不存在并发行为,因为他们对于当前会话都是独立的。
创建临时表时,ORACLE只创建了表的结构(在数据字典中定义),并没有初始化内存空间,当某一会话使用临时表时,ORALCE会
基于Nginx XSendfile+SpringMVC进行文件下载
denger
应用服务器Webnginx网络应用lighttpd
在平常我们实现文件下载通常是通过普通 read-write方式,如下代码所示。
@RequestMapping("/courseware/{id}")
public void download(@PathVariable("id") String courseID, HttpServletResp
scanf接受char类型的字符
dcj3sjt126com
c
/*
2013年3月11日22:35:54
目的:学习char只接受一个字符
*/
# include <stdio.h>
int main(void)
{
int i;
char ch;
scanf("%d", &i);
printf("i = %d\n", i);
scanf("%
学编程的价值
dcj3sjt126com
编程
发一个人会编程, 想想以后可以教儿女, 是多么美好的事啊, 不管儿女将来从事什么样的职业, 教一教, 对他思维的开拓大有帮助
像这位朋友学习:
http://blog.sina.com.cn/s/articlelist_2584320772_0_1.html
VirtualGS教程 (By @林泰前): 几十年的老程序员,资深的
二维数组(矩阵)对角线输出
飞天奔月
二维数组
今天在BBS里面看到这样的面试题目,
1,二维数组(N*N),沿对角线方向,从右上角打印到左下角如N=4: 4*4二维数组
{ 1 2 3 4 }
{ 5 6 7 8 }
{ 9 10 11 12 }
{13 14 15 16 }
打印顺序
4
3 8
2 7 12
1 6 11 16
5 10 15
9 14
13
要
Ehcache(08)——可阻塞的Cache——BlockingCache
234390216
并发ehcacheBlockingCache阻塞
可阻塞的Cache—BlockingCache
在上一节我们提到了显示使用Ehcache锁的问题,其实我们还可以隐式的来使用Ehcache的锁,那就是通过BlockingCache。BlockingCache是Ehcache的一个封装类,可以让我们对Ehcache进行并发操作。其内部的锁机制是使用的net.
mysqldiff对数据库间进行差异比较
jackyrong
mysqld
mysqldiff该工具是官方mysql-utilities工具集的一个脚本,可以用来对比不同数据库之间的表结构,或者同个数据库间的表结构
如果在windows下,直接下载mysql-utilities安装就可以了,然后运行后,会跑到命令行下:
1) 基本用法
mysqldiff --server1=admin:12345
spring data jpa 方法中可用的关键字
lawrence.li
javaspring
spring data jpa 支持以方法名进行查询/删除/统计。
查询的关键字为find
删除的关键字为delete/remove (>=1.7.x)
统计的关键字为count (>=1.7.x)
修改需要使用@Modifying注解
@Modifying
@Query("update User u set u.firstna
Spring的ModelAndView类
nicegege
spring
项目中controller的方法跳转的到ModelAndView类,一直很好奇spring怎么实现的?
/*
* Copyright 2002-2010 the original author or authors.
*
* Licensed under the Apache License, Version 2.0 (the "License");
* yo
搭建 CentOS 6 服务器(13) - rsync、Amanda
rensanning
centos
(一)rsync
Server端
# yum install rsync
# vi /etc/xinetd.d/rsync
service rsync
{
disable = no
flags = IPv6
socket_type = stream
wait
Learn Nodejs 02
toknowme
nodejs
(1)npm是什么
npm is the package manager for node
官方网站:https://www.npmjs.com/
npm上有很多优秀的nodejs包,来解决常见的一些问题,比如用node-mysql,就可以方便通过nodejs链接到mysql,进行数据库的操作
在开发过程往往会需要用到其他的包,使用npm就可以下载这些包来供程序调用
&nb
Spring MVC 拦截器
xp9802
spring mvc
Controller层的拦截器继承于HandlerInterceptorAdapter
HandlerInterceptorAdapter.java 1 public abstract class HandlerInterceptorAdapter implements HandlerIntercep