hisat2 -t --dta -x grch38/genome -1 CRC/10samples/105TRA/105TRA_1.fq.gz -2 CRC/10samples/105TRA/105TRA_2.fq.gz -S aligned/105TRA/105TRA.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/108TRA/108TRA_1.fq.gz -2 CRC/10samples/108TRA/108TRA_2.fq.gz -S aligned/108TRA/108TRA.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/112A/112A_1.fq.gz -2 CRC/10samples/112A/112A_2.fq.gz -S aligned/112A/112A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/113A/113A_1.fq.gz -2 CRC/10samples/113A/113A_2.fq.gz -S aligned/113A/113A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/114A/114A_1.fq.gz -2 CRC/10samples/114A/114A_2.fq.gz -S aligned/114A/114A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/122A/122A_1.fq.gz -2 CRC/10samples/122A/122A_2.fq.gz -S aligned/122A/122A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/124A/124A_1.fq.gz -2 CRC/10samples/124A/124A_2.fq.gz -S aligned/124A/124A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/126A/126A_1.fq.gz -2 CRC/10samples/126A/126A_2.fq.gz -S aligned/126A/126A.sam
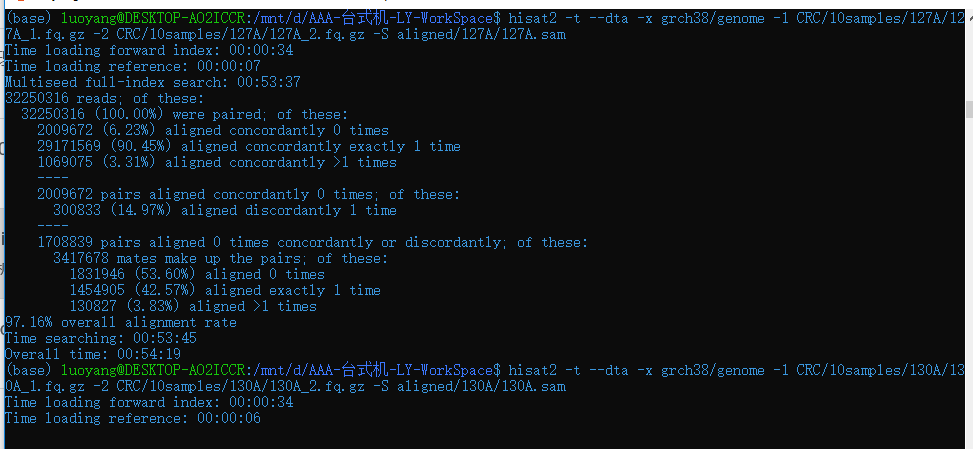
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/127A/127A_1.fq.gz -2 CRC/10samples/127A/127A_2.fq.gz -S aligned/127A/127A.sam
hisat2 -t --dta -x grch38/genome -1 CRC/10samples/130A/130A_1.fq.gz -2 CRC/10samples/130A/130A_2.fq.gz -S aligned/130A/130A.sam
忘记了创建114A的文件夹,导致该命令报错,下次记得检查一下!!!!
最后一条命令在控制台中没有运行,下次应在后面随便添加一条命令!!!!
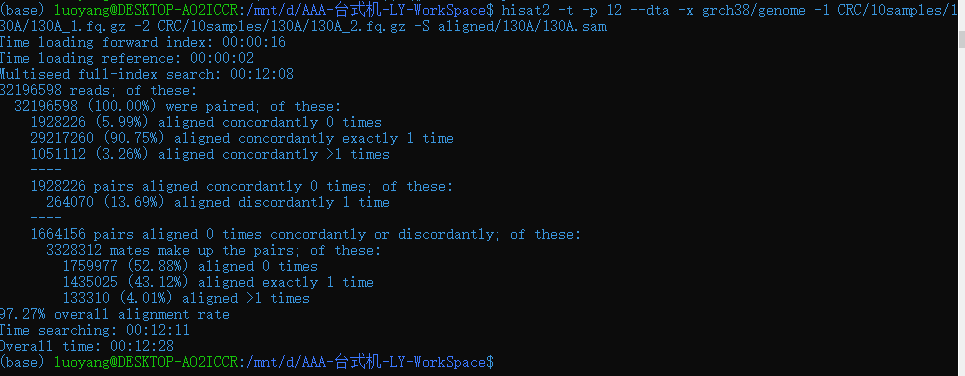
-p 设置线程数
此电脑
所以线程数最高可设置12
与单线程相比时间是之前的1/5
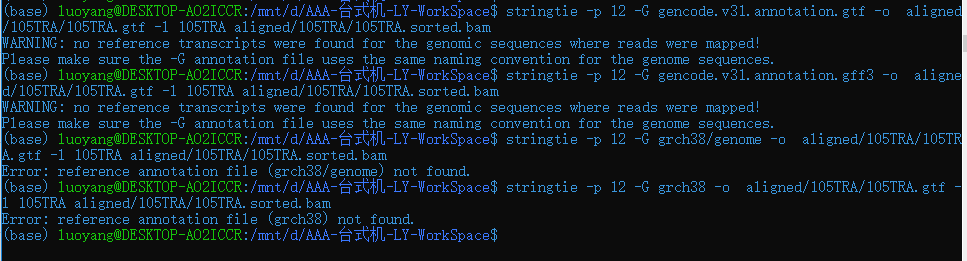
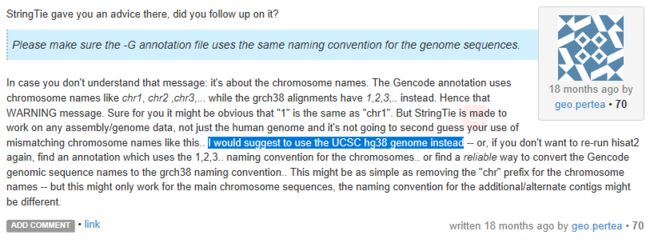
但是在后续stringtie处理过程中报错
网上查原因可能是基因命名方式不同(?)
所以又在Hisat网站上重新下载注释基因,第一次下载的是红色的,这次下载黄色方框试一试看(貌似没报错)
Align the RNA-seq reads to the genome
第一步:Map the reads for each sample to the reference genome
(base) luoyang@DESKTOP-AO2ICCR:/mnt/d/AAA-台式机-LY-WorkSpace$ hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/130A/130A_1.fq.gz -2 CRC/10samples/130A/130A_2.fq.gz -S aligned/130A/130A.sam
Time loading forward index: 00:00:38
Time loading reference: 00:00:07
Multiseed full-index search: 00:12:35
32196598 reads; of these:
32196598 (100.00%) were paired; of these:
2022163 (6.28%) aligned concordantly 0 times
28316980 (87.95%) aligned concordantly exactly 1 time
1857455 (5.77%) aligned concordantly >1 times
----
2022163 pairs aligned concordantly 0 times; of these:
250312 (12.38%) aligned discordantly 1 time
----
1771851 pairs aligned 0 times concordantly or discordantly; of these:
3543702 mates make up the pairs; of these:
1847559 (52.14%) aligned 0 times
1402980 (39.59%) aligned exactly 1 time
293163 (8.27%) aligned >1 times
97.13% overall alignment rate
Time searching: 00:12:43
Overall time: 00:13:22
最后生成105TRA.sam文件
新命令:(10samples)
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/105TRA/105TRA_1.fq.gz -2 CRC/10samples/105TRA/105TRA_2.fq.gz -S aligned/105TRA/105TRA.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/108TRA/108TRA_1.fq.gz -2 CRC/10samples/108TRA/108TRA_2.fq.gz -S aligned/108TRA/108TRA.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/112A/112A_1.fq.gz -2 CRC/10samples/112A/112A_2.fq.gz -S aligned/112A/112A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/113A/113A_1.fq.gz -2 CRC/10samples/113A/113A_2.fq.gz -S aligned/113A/113A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/114A/114A_1.fq.gz -2 CRC/10samples/114A/114A_2.fq.gz -S aligned/114A/114A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/122A/122A_1.fq.gz -2 CRC/10samples/122A/122A_2.fq.gz -S aligned/122A/122A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/124A/124A_1.fq.gz -2 CRC/10samples/124A/124A_2.fq.gz -S aligned/124A/124A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/126A/126A_1.fq.gz -2 CRC/10samples/126A/126A_2.fq.gz -S aligned/126A/126A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/127A/127A_1.fq.gz -2 CRC/10samples/127A/127A_2.fq.gz -S aligned/127A/127A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/10samples/130A/130A_1.fq.gz -2 CRC/10samples/130A/130A_2.fq.gz -S aligned/130A/130A.sam
16samples:
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/17F24T/17F24T_1.fq.gz -2 CRC/16samples/17F24T/17F24T_2.fq.gz -S aligned-16/17F24T/17F24T.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/105TRA/105TRA_1.fq.gz -2 CRC/16samples/105TRA/105TRA_2.fq.gz -S aligned-16/105TRA/105TRA.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/109TRA/109TRA_1.fq.gz -2 CRC/16samples/109TRA/109TRA_2.fq.gz -S aligned-16/109TRA/109TRA.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/110A/110A_1.fq.gz -2 CRC/16samples/110A/110A_2.fq.gz -S aligned-16/110A/110A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/113A/113A_1.fq.gz -2 CRC/16samples/113A/113A_2.fq.gz -S aligned-16/113A/113A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/114A/114A_1.fq.gz -2 CRC/16samples/114A/114A_2.fq.gz -S aligned-16/114A/114A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/120A/120A_1.fq.gz -2 CRC/16samples/120A/120A_2.fq.gz -S aligned-16/120A/120A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/121A/121A_1.fq.gz -2 CRC/16samples/121A/121A_2.fq.gz -S aligned-16/121A/121A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/122A/122A_1.fq.gz -2 CRC/16samples/122A/122A_2.fq.gz -S aligned-16/122A/122A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/124A/124A_1.fq.gz -2 CRC/16samples/124A/124A_2.fq.gz -S aligned-16/124A/124A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/126A/126A_1.fq.gz -2 CRC/16samples/126A/126A_2.fq.gz -S aligned-16/126A/126A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/136A/136A_1.fq.gz -2 CRC/16samples/136A/136A_2.fq.gz -S aligned-16/136A/136A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/140A/140A_1.fq.gz -2 CRC/16samples/140A/140A_2.fq.gz -S aligned-16/140A/140A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/143A/143A_1.fq.gz -2 CRC/16samples/143A/143A_2.fq.gz -S aligned-16/143A/143A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/153A/153A_1.fq.gz -2 CRC/16samples/153A/153A_2.fq.gz -S aligned-16/153A/153A.sam
hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/154A/154A_1.fq.gz -2 CRC/16samples/154A/154A_2.fq.gz -S aligned-16/154A/154A.sam
其中121A报错:后来发现是该文件夹中没有121A_2.fq.gz,加上后就正常了
(base) luoyang@DESKTOP-AO2ICCR:/mnt/d/AAA-台式机-LY-WorkSpace$ hisat2 -t -p 12 --dta -x hg38/genome -1 CRC/16samples/121A/121A_1.fq.gz -2 CRC/16samples/121A/121A_2.fq.gz -S aligned-16/121A/121A.sam
gzip: CRC/16samples/121A/121A_2.fq.gz: No such file or directory
Time loading forward index: 00:00:38
Time loading reference: 00:00:07
Error, fewer reads in file specified with -2 than in file specified with -1
terminate called after throwing an instance of 'int'
Aborted (core dumped)
(ERR): hisat2-align exited with value 134