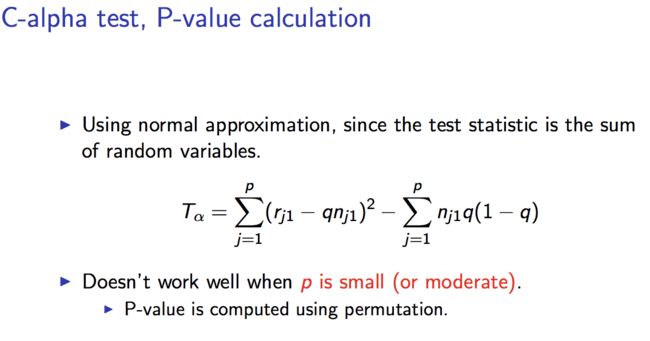
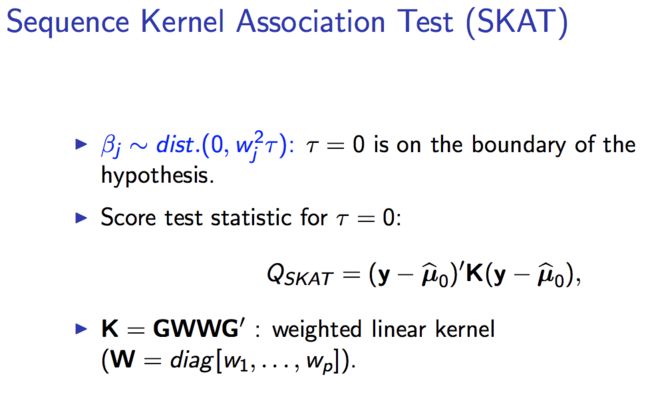
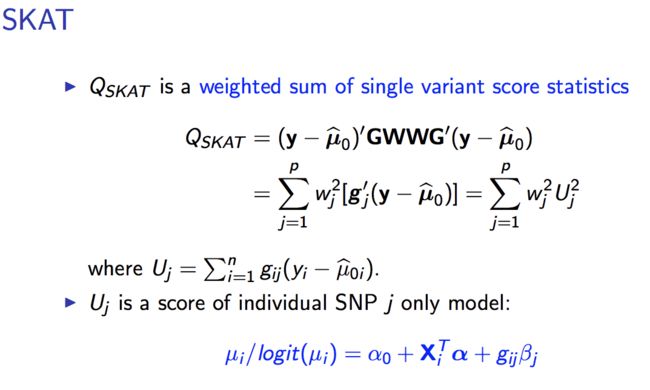
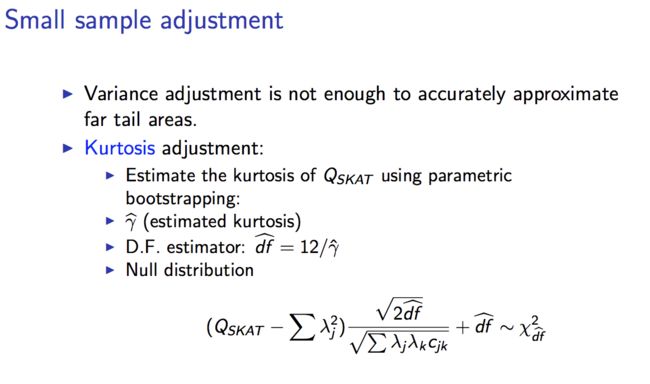
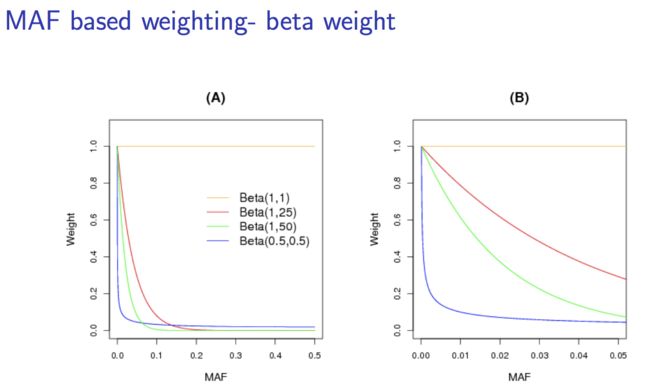
-
p 越小,效力越大
##################################################################################################################
### R Scripts: let's read Gene2 into R and analyze it
library(SKAT)
Z = as.matrix(read.table("Gene2.txt", header = F))
maf = apply(Z,2,mean)/2 ## calculate the mean of each column and divide by 2 to get the maf
y.c = scan("Trait2.txt") ## read in the values for trait 1
rvs = which(maf<0.03) ## Set the threshold to be 0.03
obj<-SKAT_Null_Model(y.c ~ 1, out_type="C") ## calculates the NULL model, i.e. there are no variants
SKAT(Z, obj)$p.value ## test for trait 2 with gene 2. Note that we are using the default settings and testing all variants.
SKAT(Z[,rvs], obj)$p.value ## restrict attention to rare variants. The null model has not changed.
- 定性性状
##################################################################################################################
##################################################################################################################
### R Scripts: let's dichtomize trait 2
q1 = quantile(y.c,0.25)
q3 = quantile(y.c,0.75)
y.b = rep(NA, length(y.c))
y.b[which(y.c <= q1)] = 0
y.b[which(y.c >= q3)] = 1
omits = which(is.na(y.b))
y.b = y.b[-omits]
Z.b = Z[-omits,]
obj<-SKAT_Null_Model(y.b ~ 1, out_type="D") ## calculates the NULL model, i.e. there are no variants
SKAT(Z.b, obj)$p.value ## test for trait 2 with gene 2. Note that we are using the default settings and testing all variants.
SKAT(Z.b[,rvs], obj)$p.value ## restrict attention to rare variants. The null model has not changed.
-
gene2.txt 1000个人,62个位点
##################################################################################################################
##################################################################################################################
### R Scripts: let's try running the C-alpha; recall that this can be done by setting the weights to be 1!
obj<-SKAT_Null_Model(y.c ~ 1, out_type="C") ## calculates the NULL model, i.e. there are no variants -- need to rerun because outcome changed
SKAT(Z[,rvs], obj, weights.beta = c(1,1))$p.value ## restrict attention to rare variants. The null model has not changed.
##################################################################################################################
##################################################################################################################
### R Scripts: omnibus version of SKAT with several different rho values
SKAT(Z[,rvs], obj, r.corr=0)$p.value ## same as running SKAT
SKAT(Z[,rvs], obj, r.corr=1)$p.value ## same as weighted count collapsing
SKAT(Z[,rvs], obj, r.corr=.5)$p.value ## something in between
weights = dbeta(maf[rvs], 1,25)
C = Z[,rvs]%*%weights ## calculate the collapsed variable
summary(lm(y.c~C))
##################################################################################################################
##################################################################################################################
### R Scripts: omnibus version of SKAT with "optimal" rho value
SKAT(Z[,rvs], obj, method="optimal")$p.value
SKAT(Z[,rvs], obj, method="optimal.adj")$p.value ## slightly better type I error control in the tails.
##################################################################################################################
##################################################################################################################
### R Scripts: read in PLINK example data and analyze the 10 genes there.
## define the file names and paths, note that the SSD and SSD.info don't exist yet
File.Bed<-"Example1.bed"
File.Bim<-"Example1.bim"
File.Fam<-"Example1.fam"
File.SetID<-"Example1.SetID"
File.SSD<-"Example1.SSD"
File.Info<-"Example1.SSD.info"
## To use binary ped files, you have to generate SSD file first.
## If you already have a SSD file, you do not need to call this
Generate_SSD_SetID(File.Bed, File.Bim, File.Fam, File.SetID, File.SSD, File.Info)
## Now we can open the SSD files and also run SKAT
FAM<-Read_Plink_FAM(File.Fam, Is.binary=FALSE)
y<-FAM$Phenotype
## To use a SSD file, please open it first.
## After finishing using it, you must close it.
SSD.INFO<-Open_SSD(File.SSD, File.Info)
## Number of samples
SSD.INFO$nSample
## Number of Sets
SSD.INFO$nSets
obj<-SKAT_Null_Model(y ~ 1, out_type="C")
out<-SKAT.SSD.All(SSD.INFO, obj)
out