上一节数量生态学笔记||冗余分析(RDA)概述中,我们回顾了RDA的计算过程,不管这个过程我们有没有理解透彻,我希望你能知道的是:RDA是响应变量矩阵与解释变量之间多元多重线性回归的拟合值矩阵的PCA分析。本节我们就是具体来看一个RDA的分析案例,来看看里面的参数以及结果的解读。
# CHAPTER 6 - CANONICAL ORDINATION
# ********************************
# 载入所需程序包
library(ade4)
library(vegan)
library(packfor)# 可在http://r-forge.r-project.org/R/?group_id=195下载,但是好像在R 3.5.1上加载不了,所以这篇我用R3.4来做的。packfor已经不用,函数都搬到adespatial
# 如果是MacOS X系统,packfor程序包内forward.sel函数的运行需要加载
# gfortran程序包。用户必须从"cran.r-project.org"网站内选择"MacOS X",
# 然后选择"tools"安装gfortran程序包。
rm(list = ls())
setwd("D:\\Users\\Administrator\\Desktop\\RStudio\\数量生态学\\DATA")
library(MASS)
library(ellipse)
library(FactoMineR)
# 附加函数
source("evplot.R")
source("hcoplot.R")
# 导入CSV数据文件
spe <- read.csv("DoubsSpe.csv", row.names=1)
env <- read.csv("DoubsEnv.csv", row.names=1)
spa <- read.csv("DoubsSpa.csv", row.names=1)
# 删除没有数据的样方8
spe <- spe[-8, ]
env <- env[-8, ]
spa <- spa[-8, ]
# 提取环境变量das(离源头距离)以备用
das <- env[, 1]
# 从环境变量矩阵剔除das变量
env <- env[, -1]
# 将slope变量(pen)转化为因子(定性)变量
pen2 <- rep("very_steep", nrow(env))
pen2[env$pen <= quantile(env$pen)[4]] = "steep"
pen2[env$pen <= quantile(env$pen)[3]] = "moderate"
pen2[env$pen <= quantile(env$pen)[2]] = "low"
pen2 <- factor(pen2, levels=c("low", "moderate", "steep", "very_steep"))
table(pen2)
# 生成一个含定性坡度变量的环境变量数据框env2
env2 <- env
env2$pen <- pen2
# 将所有解释变量分为两个解释变量子集
# 地形变量(上下游梯度)子集
envtopo <- env[, c(1:3)]
names(envtopo)
#水体化学属性变量子集
envchem <- env[, c(4:10)]
names(envchem)
# 物种数据Hellinger转化
spe.hel <- decostand(spe, "hellinger")
使用vegan包运行RDA
vegan包运行RDA有两种不同的模式。第一种是简单模式,直接输入用逗号隔开的数据矩阵对象到rda()函数:
式中为响应变量矩阵,为解释变量矩阵,为偏RDA分析需要的协变量矩阵。
此公式有一个缺点:不能有因子变量(定性变量)。如果有因子变量,建议使用第二种模式:
式中,为响应变量矩阵。解释变量矩阵包括定量变量(var1)、因子变量(factorA)以及变量2和变量3的交互作用项,协变量(var4)被放到Condition()里。所用的数据都放在XWdata的数据框里。
这个公式与lm()函数以及其他回归函数一样,左边是响应变量,右边是解释变量。
# 基于Hellinger 转化的鱼类数据RDA,解释变量为对象env2包括的环境变量
# 关注省略模式的公式
spe.rda <- rda(spe.hel~., env2)
summary(spe.rda) # 2型标尺(默认)
#这里使用一些默认的选项,即 scale=FALSE(基于协方差矩阵的RDA)和#scaling=2
RDA结果的摘录:
RDA formula :
Call:
rda(formula = spe.hel ~ alt + pen + deb + pH +
dur + pho + nit + amm + oxy + dbo, data = env2)
方差分解(Partitioning of variance):总方差被划分为约束和非约束两部分。约束部分表示响应变量矩阵的总方差能被解释变量解释的部分,如果用比例来表示,其值相当于多元回归的。在RDA中,这个解释比例值也称作双多元冗余统计。然而,类似多元回归的未校正的,RDA的是有偏差的,需要进一步校正。
Partitioning of variance:
Inertia Proportion
Total 0.5025 1.0000
Constrained 0.3654 0.7271
Unconstrained 0.1371 0.2729
特征根以及对方差的贡献率(Eigenvalues, and their contribution to the variance ):当前这个RDA分析产生了12个典范轴(特征根用RDA1 至RDA12表示)和16个非约束轴(特征根用PC1至PC16表示)。输出结果不仅包含每轴特征根同时也给出累积方差解释率(约束轴)或承载轴(非约束轴),最终的累计值必定是1.12 个典范轴累积解释率也代表响应变量总方差能够被解释变量解释的部分。
两个特征根的重要区别:典范特征根RDAx是响应变量总方差能够被解释变量解释的部分,而残差特征根RCx响应变量总方差能够被残差轴解释的部分,与RDA无关。
Eigenvalues, and their contribution to the variance
Importance of components:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11 RDA12 PC1 PC2 PC3
Eigenvalue 0.2281 0.0537 0.03212 0.02321 0.008707 0.007218 0.004862 0.002919 0.002141 0.001160 0.0009134 0.0003406 0.04580 0.02814 0.01529
Proportion Explained 0.4539 0.1069 0.06393 0.04618 0.017328 0.014363 0.009676 0.005809 0.004260 0.002308 0.0018176 0.0006778 0.09115 0.05600 0.03042
Cumulative Proportion 0.4539 0.5607 0.62467 0.67085 0.688176 0.702539 0.712215 0.718025 0.722284 0.724592 0.7264100 0.7270878 0.81824 0.87424 0.90466
PC4 PC5 PC6 PC7 PC8 PC9 PC10 PC11 PC12 PC13 PC14 PC15 PC16
Eigenvalue 0.01399 0.009841 0.007676 0.004206 0.003308 0.002761 0.002016 0.001752 0.0009851 0.0005921 0.0004674 0.0002127 0.0001004
Proportion Explained 0.02784 0.019584 0.015276 0.008371 0.006583 0.005495 0.004013 0.003486 0.0019604 0.0011783 0.0009301 0.0004233 0.0001998
Cumulative Proportion 0.93250 0.952084 0.967360 0.975731 0.982314 0.987809 0.991822 0.995308 0.9972684 0.9984468 0.9993768 0.9998002 1.0000000
累积约束特征根(Accumulated constrained eigenvalues)表示在本轴以及前面所有轴的典范轴所能解释的方差占全部解释方差的比例累积。
Accumulated constrained eigenvalues
Importance of components:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10 RDA11 RDA12
Eigenvalue 0.2281 0.0537 0.03212 0.02321 0.008707 0.007218 0.004862 0.002919 0.002141 0.001160 0.0009134 0.0003406
Proportion Explained 0.6242 0.1470 0.08792 0.06352 0.023832 0.019755 0.013308 0.007990 0.005859 0.003174 0.0024999 0.0009322
Cumulative Proportion 0.6242 0.7712 0.85913 0.92265 0.946483 0.966237 0.979545 0.987535 0.993394 0.996568 0.9990678 1.0000000
物种得分(Species scores)双序图和三序图内代表响应变量的箭头的顶点坐标。与PCA相同,坐标依赖标尺Scaling的选择。
Scaling 2 for species and site scores
* Species are scaled proportional to eigenvalues
* Sites are unscaled: weighted dispersion equal on all dimensions
* General scaling constant of scores: 1.93676
Species scores
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6
CHA 0.13383 0.11623 -0.238180 0.018611 0.043221 -0.029737
TRU 0.64238 0.06648 0.123713 0.181572 -0.009691 0.029793
VAI 0.47475 0.07015 -0.010218 -0.115369 -0.045317 -0.030033
LOC 0.36260 0.06972 0.041240 -0.190586 -0.046881 0.006448
OMB 0.13079 0.10709 -0.239224 0.043603 0.065881 0.003458
BLA 0.06587 0.12475 -0.216900 -0.004157 0.021793 -0.004195
HOT -0.17417 0.06778 -0.008426 -0.016419 -0.079730 0.044706
TOX -0.12683 0.16052 -0.035733 -0.016087 -0.089768 -0.001880
VAN -0.07963 0.04200 0.007636 -0.059179 -0.033596 -0.121440
CHE -0.10903 -0.17552 -0.090099 -0.168373 0.019444 0.008745
BAR -0.18528 0.21154 -0.073087 -0.006879 -0.012995 0.040484
SPI -0.16064 0.15513 -0.014309 -0.002488 -0.060810 0.011045
GOU -0.20537 0.02484 -0.007973 -0.017742 -0.049137 -0.096231
BRO -0.10734 0.02848 0.090055 0.012324 0.075184 -0.057088
PER -0.09164 0.10506 0.070393 -0.057443 0.013870 -0.009906
BOU -0.20907 0.16002 0.025500 0.012078 -0.011477 0.022035
PSO -0.22799 0.11121 0.018800 -0.009474 -0.027431 0.024517
ROT -0.16098 0.01348 0.041628 0.032398 0.054117 -0.094582
CAR -0.17384 0.14901 0.022262 0.009534 0.004991 -0.005396
TAN -0.14025 0.10632 0.078290 -0.122627 0.054162 0.031256
BCO -0.18594 0.12222 0.053881 0.026170 0.044015 0.014577
PCH -0.14630 0.08894 0.061880 0.034763 0.083530 0.004396
GRE -0.30881 0.01606 0.039366 0.029254 -0.011141 -0.052412
GAR -0.31982 -0.16601 -0.018225 -0.115454 0.054341 0.064772
BBO -0.23897 0.09090 0.051627 0.010224 0.007004 0.036497
ABL -0.43215 -0.22639 -0.108190 0.138807 -0.083920 0.008460
ANG -0.19442 0.14149 0.033659 0.017387 0.008110 0.017638
样方得分(Site scores (weighted sums of species scores))物种得分的加权和:使用响应变量矩阵计算获得的样方坐标。
Site scores (weighted sums of species scores)
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6
1 0.40151 -0.154306 0.55539 1.600773 0.191866 0.916893
2 0.53523 -0.025084 0.43389 0.294615 -0.518456 0.458860
3 0.49430 -0.014605 0.49409 0.169038 -0.246166 0.163421
4 0.33452 0.001173 0.51626 -0.321009 0.088716 -0.219837
5 0.02794 -0.194357 0.44612 -0.559210 0.853768 -1.115654
6 0.24422 -0.130778 0.41372 -0.696264 0.181514 -0.273473
7 0.46590 -0.125982 0.31674 -0.137834 -0.548635 -0.061703
9 0.03662 -0.605060 -0.07022 -1.260916 0.669108 1.164986
10 0.31381 -0.198654 0.10764 -0.635139 -0.741448 -0.990236
11 0.48116 -0.039598 -0.37851 0.181924 0.221494 0.254511
12 0.49162 0.014263 -0.37983 0.163103 0.223730 0.324672
13 0.49848 0.212367 -0.67408 0.518823 0.400091 0.221622
14 0.38202 0.229538 -0.75771 0.223651 0.515712 -0.139740
15 0.28739 0.218713 -0.71887 -0.210821 0.176392 -0.553185
16 0.09129 0.400192 -0.34443 -0.376097 -0.366868 -0.575230
17 -0.05306 0.423994 -0.41009 -0.188492 -0.726152 0.151876
18 -0.14185 0.385926 -0.36814 -0.217143 -0.644298 -0.001052
19 -0.28204 0.275528 -0.01877 -0.371457 -0.691725 -0.062230
20 -0.39683 0.209468 0.11547 -0.177972 -0.387121 0.048690
21 -0.42851 0.278256 0.22010 -0.005993 -0.027083 -0.042209
22 -0.46553 0.251819 0.22784 0.040192 0.152965 0.032185
23 -0.28123 -1.145599 -0.50543 0.300015 -0.004403 1.157206
24 -0.40893 -0.752909 -0.26785 0.428851 -0.645606 0.643084
25 -0.35205 -0.770380 -0.12186 0.459170 0.078892 -1.725973
26 -0.46923 0.093958 0.23058 0.107865 0.310432 0.132556
27 -0.47021 0.213521 0.24887 0.084219 0.331685 0.125439
28 -0.47266 0.233922 0.27053 0.105776 0.381436 0.093719
29 -0.37457 0.393260 0.10423 0.202115 0.282621 0.021834
30 -0.48932 0.321417 0.31431 0.278218 0.487541 -0.151031
样方约束——解释变量的线性组合(Site constraints (linear combinations of constraining variables)):使用解释变量矩阵计算获得的样方坐标,是拟合的(fitted)样方坐标。
Site constraints (linear combinations of constraining variables)
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6
1 0.55135 0.002395 0.47774 0.626878 -0.210700 0.31511
2 0.29737 0.105715 0.64862 0.261161 -0.057741 0.09322
3 0.36834 -0.185376 0.59788 0.324212 -0.002385 0.31090
4 0.44348 -0.066086 0.33260 -0.344463 -0.279591 -0.37079
5 0.27004 -0.366721 0.17992 -0.453274 0.716614 -0.06545
6 0.37148 -0.255624 0.40847 0.217259 0.023374 0.34360
7 0.53874 -0.179999 0.06845 -0.424896 0.024884 -0.33454
9 -0.04438 -0.362632 0.12371 -1.180662 0.348188 0.26352
10 0.16289 -0.154212 0.22252 -0.241425 -0.573048 -0.02867
11 0.29912 0.176150 -0.08233 0.003924 0.164806 -0.44603
12 0.36843 0.197492 -0.41095 0.300566 -0.053922 -0.01139
13 0.42626 0.190107 -0.59764 0.100988 0.118714 -0.21022
14 0.34373 0.134362 -0.80378 0.063879 0.665797 0.48126
15 0.21385 0.237182 -0.56341 -0.001099 -0.028564 0.01655
16 0.03056 0.352192 -0.12110 -0.202316 0.058413 -0.43542
17 -0.10499 0.178587 -0.26925 0.046988 -0.608314 0.21237
18 -0.11204 0.221631 -0.24024 -0.302957 -0.251346 -0.01448
19 -0.05479 0.311860 -0.30701 0.010366 -0.481829 -0.12855
20 -0.25684 0.303770 -0.06768 -0.036587 -0.562578 0.13698
21 -0.39177 0.196355 0.01877 -0.281086 -0.383524 0.39310
22 -0.21361 0.180414 0.01066 0.074301 -0.036849 -0.02429
23 -0.21654 -1.016853 -0.57298 0.548175 0.182594 0.51443
24 -0.52578 -0.645438 -0.11182 -0.240149 -0.512492 0.32420
25 -0.38886 -0.867381 -0.08079 0.482839 -0.106743 -1.21305
26 -0.48440 0.031510 0.14065 -0.114545 0.425712 -0.17989
27 -0.61221 0.138191 0.32316 -0.015795 0.232397 0.38288
28 -0.46921 0.459843 0.22002 0.078870 0.278747 -0.36504
29 -0.38450 0.344487 0.20622 0.353008 0.504693 0.17747
30 -0.42572 0.338080 0.24960 0.345839 0.404692 -0.13777
解释变量双序图得分(Biplot scores for constraining variables):排序图内解释解释变量箭头的坐标,按照下面的过程获得:运行解释变量与拟合的样方坐标之间的相关分析,然后将所有相关系数转化为双序图内坐标。所有的变量包括个水平的因子口可以有自己的坐标对因子变量在排序轴的坐标,用各个因子的形心表示更合适。
Biplot scores for constraining variables
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6
alt 0.8239 -0.203257 0.46604 -0.16936 0.003229 0.10407
penmoderate -0.3592 -0.008729 -0.21727 -0.18278 0.157934 0.50094
pensteep 0.2791 0.156027 -0.06876 0.01927 0.176390 -0.15469
penvery_steep 0.6129 -0.148496 0.45379 0.03618 -0.191046 -0.04715
deb -0.7770 0.254952 -0.17470 0.30995 0.227580 -0.11938
pH 0.1023 0.178431 -0.30131 0.03959 0.298584 0.04848
dur -0.5722 0.044963 -0.56040 -0.14813 0.275617 -0.24527
pho -0.4930 -0.650488 -0.19868 0.29286 0.055893 -0.39345
nit -0.7755 -0.203836 -0.23285 0.19744 -0.078849 -0.35073
amm -0.4744 -0.687577 -0.16648 0.28470 -0.051768 -0.33852
oxy 0.7632 0.575528 -0.16425 0.08026 -0.136143 0.13748
dbo -0.5171 -0.791805 -0.15652 0.22064 0.075568 -0.09105
因子解释变量形心(Centroids for factor constraints):因子变量各个水平形心点的坐标,即每个水平所用标识为一的样方的形心。
Centroids for factor constraints
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6
penlow -0.2800 0.005530 -0.09025 0.07614 -0.07860 -0.18390
penmoderate -0.2093 -0.005086 -0.12660 -0.10650 0.09203 0.29189
pensteep 0.1965 0.109867 -0.04842 0.01357 0.12420 -0.10892
penvery_steep 0.3908 -0.094679 0.28933 0.02307 -0.12181 -0.03006
在rda()函数中大家感兴趣的典范特征系数(即每个解释变量与每个典范轴之间的回归系数),可用coef()函数获得:
#如何从rda()输出结果中获得典范系数
coef(spe.rda)
alt 0.0004482548 7.559499e-05 0.0005205825 0.0003883845 0.001857206 -6.313946e-05 -0.001355362 0.001120849 -0.0002530083 0.001189659
penmoderate -0.0123961693 -1.660194e-02 0.0160069104 -0.0278562534 0.276128846 1.310695e-01 -0.022918427 0.018830063 -0.3113354204 -0.278006278
pensteep 0.0478390238 4.893302e-02 0.1022577908 0.1347997771 0.393812929 -1.795824e-01 0.046319741 0.123642821 0.0963820046 -0.447099975
penvery_steep 0.0180005587 -5.691933e-02 0.2322637617 0.1002359565 0.036814635 -1.742060e-01 0.517299284 0.067564014 -0.2262450630 -0.590022907
deb -0.0014063766 4.440084e-03 0.0089298486 0.0164715901 0.013318449 2.705757e-03 -0.002419805 0.010394632 -0.0006430624 0.004427139
pH 0.0188227657 -3.163592e-02 -0.0482021704 0.1141647498 0.412886847 1.091403e-01 0.139806409 -0.436295510 -0.0215841003 -0.904063764
dur 0.0025580344 -1.955496e-03 -0.0065901935 -0.0093556696 0.005228707 -6.098098e-03 0.002195518 0.010536248 -0.0006844877 0.003353110
pho 0.1031541920 4.583584e-02 -0.1000153096 -0.1050243435 0.422991234 -3.694132e-01 0.035874664 -0.701043138 -0.2865315085 0.245917738
nit -0.0123824749 1.041485e-01 0.0625396719 0.0774218297 0.234401221 -3.541252e-02 -0.240428544 0.128403162 -0.0686364968 0.113090041
amm -0.1084411839 -4.407786e-01 0.0057247742 0.0538542716 -1.812468883 4.798631e-03 0.281937862 1.068013480 0.3084215704 -1.217501183
oxy 0.0686692124 1.980446e-02 -0.0894153251 0.1200884061 0.032052566 3.880445e-02 -0.058026043 0.061374900 -0.0196444146 0.089881042
dbo 0.0108322463 -2.696114e-02 -0.0253225230 0.0745175780 0.067082880 9.276548e-02 -0.019719504 0.047865971 0.0359365102 0.065416035
RDA11 RDA12
alt 0.0006826822 0.0009471677
penmoderate 0.0398080898 -0.2974896027
pensteep 0.2445035939 -0.3475535793
penvery_steep 0.2457103975 -0.1848717482
deb -0.0022565029 0.0064858596
pH 0.0696045266 0.5756301035
dur 0.0007758175 0.0062181193
pho -0.0015544897 -0.6309008260
nit 0.3983543655 0.0942246573
amm -1.5964107514 0.8979015239
oxy 0.0627415675 0.0258937429
dbo 0.1113928484 0.0403158432
提取。解读和绘制vegan包输出的RDA结果
校正
# 提取校正R2
# **********
# 从rda的结果中提取未校正R2
(R2 <- RsquareAdj(spe.rda)$r.squared)
[1] 0.7270878
# 从rda的结果中提取校正R2
(R2adj <- RsquareAdj(spe.rda)$adj.r.squared)
[1] 0.5224036
#可以看出,校正R2总是小于R2。校正R2作为被解释方差比例的无偏估计,后#面的变差分解部分所用的也是校正R2。
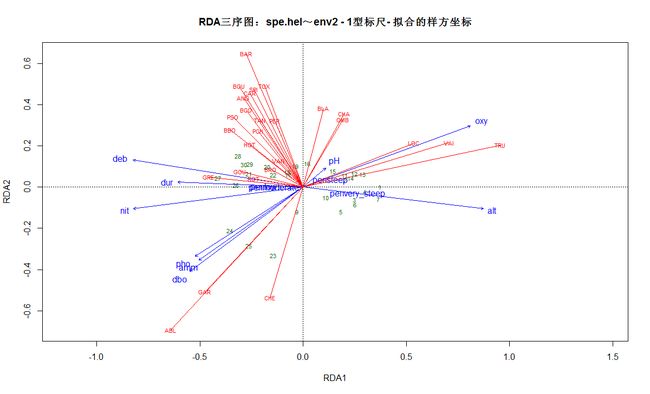
# RDA三序图
现在绘制RDA的排序图。如果一张排序图中有三个实体:样方、响应变量、解释变量,这种排序图称为三序图(triplot)为了区分响应变量和解释变量,定量解释变量用箭头表示,响应变量用不带箭头的线表示。
# RDA三序图
# **********
# 1型标尺:距离三序图
plot(spe.rda, scaling=1, main="RDA三序图:spe.hel~env2 - 1型标尺- 加权和样方坐标")
#此排序图同时显示所有的元素:样方、物种、定量解释变量(用箭头表示)
#和因子变量的形心。为了与定量解释变量区分,物种用不带箭头的线表示。
spe.sc <- scores(spe.rda, choices=1:2, scaling=1, display="sp")
arrows(0, 0, spe.sc[, 1], spe.sc[, 2], length=0, lty=1, col="red")
plot(spe.rda, main="RDA三序图:spe.hel~env2 - 2型标尺- 加权和样方坐标")
spe2.sc <- scores(spe.rda, choices=1:2, display="sp")
arrows(0, 0, spe2.sc[, 1], spe2.sc[, 2], length=0, lty=1, col="red")
# 样方坐标是环境因子线性组合
# 1型标尺
plot(spe.rda, scaling=1, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~env2 - 1型标尺- 拟合的样方坐标")
arrows(0, 0, spe.sc[, 1], spe.sc[, 2], length=0, lty=1, col="red")
# 2型标尺
plot(spe.rda, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~env2 - 2型标尺- 拟合的样方坐标")
arrows(0, 0, spe2.sc[,1], spe2.sc[,2], length=0, lty=1, col="red")
RDA 结果的置换检验
# RDA所有轴置换检验
anova.cca(spe.rda, step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ alt + pen + deb + pH + dur + pho + nit + amm + oxy + dbo, data = env2)
Df Variance F Pr(>F)
Model 12 0.36537 3.5522 0.001 ***
Residual 16 0.13714
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
# 每个典范轴逐一检验
anova.cca(spe.rda, by="axis", step=1000)
Permutation test for rda under reduced model
Forward tests for axes
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ alt + pen + deb + pH + dur + pho + nit + amm + oxy + dbo, data = env2)
Df Variance F Pr(>F)
RDA1 1 0.228081 26.6098 0.001 ***
RDA2 1 0.053696 6.2646 0.003 **
RDA3 1 0.032124 3.7478 0.361
RDA4 1 0.023207 2.7075 0.763
RDA5 1 0.008707 1.0159 1.000
RDA6 1 0.007218 0.8421 1.000
RDA7 1 0.004862 0.5673 1.000
RDA8 1 0.002919 0.3406 1.000
RDA9 1 0.002141 0.2497 1.000
RDA10 1 0.001160 0.1353 1.000
RDA11 1 0.000913 0.1066 1.000
RDA12 1 0.000341 0.0397 1.000
Residual 16 0.137141
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#每个典范轴的检验只能输入由公式模式获得的rda结果。有多少个轴结果是
#显著的?
# 使用Kaiser-Guttman准则确定残差轴
spe.rda$CA$eig[spe.rda$CA$eig > mean(spe.rda$CA$eig)]
PC1 PC2 PC3 PC4 PC5
0.045802781 0.028143080 0.015288209 0.013987518 0.009841239
#很明显,还有一部分有意思的变差尚未被目前所用的这套环境变量解释。
偏RDA分析
偏RDA:固定地形变量影响后,水体化学属性的效应
# 简单模式:X和W可以是分离的定量变量表格
spechem.physio <- rda(spe.hel, envchem, envtopo)
spechem.physio
Call: rda(X = spe.hel, Y = envchem, Z = envtopo)
Inertia Proportion Rank
Total 0.5025 1.0000
Conditional 0.2087 0.4153 3
Constrained 0.1602 0.3189 7
Unconstrained 0.1336 0.2659 18
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7
0.09137 0.04591 0.00928 0.00625 0.00387 0.00214 0.00142
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
0.04642 0.02072 0.01746 0.01325 0.00974 0.00588 0.00512 0.00400
(Showed only 8 of all 18 unconstrained eigenvalues)
summary(spechem.physio)
# 公式模式;X和W必须在同一数据框内
class(env)
spechem.physio2 <- rda(spe.hel ~ pH + dur + pho + nit + amm + oxy + dbo
+ Condition(alt + pen + deb), data=env)
spechem.physio2
Call: rda(formula = spe.hel ~ pH + dur + pho + nit + amm + oxy + dbo + Condition(alt + pen + deb), data = env)
Inertia Proportion Rank
Total 0.5025 1.0000
Conditional 0.2087 0.4153 3
Constrained 0.1602 0.3189 7
Unconstrained 0.1336 0.2659 18
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7
0.09137 0.04591 0.00928 0.00625 0.00387 0.00214 0.00142
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
0.04642 0.02072 0.01746 0.01325 0.00974 0.00588 0.00512 0.00400
(Showed only 8 of all 18 unconstrained eigenvalues)
#上面两个分析的结果完全相同。
偏RDA检验(使用公式模式获得的RDA结果,以便检验每个轴)
anova.cca(spechem.physio2, step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ pH + dur + pho + nit + amm + oxy + dbo + Condition(alt + pen + deb), data = env)
Df Variance F Pr(>F)
Model 7 0.16024 3.0842 0.001 ***
Residual 18 0.13360
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
anova.cca(spechem.physio2, step=1000, by="axis")
Permutation test for rda under reduced model
Forward tests for axes
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ pH + dur + pho + nit + amm + oxy + dbo + Condition(alt + pen + deb), data = env)
Df Variance F Pr(>F)
RDA1 1 0.091373 12.3108 0.001 ***
RDA2 1 0.045907 6.1851 0.010 **
RDA3 1 0.009277 1.2499 0.964
RDA4 1 0.006251 0.8422 0.993
RDA5 1 0.003866 0.5209 0.996
RDA6 1 0.002142 0.2886 1.000
RDA7 1 0.001425 0.1920 0.997
Residual 18 0.133599
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
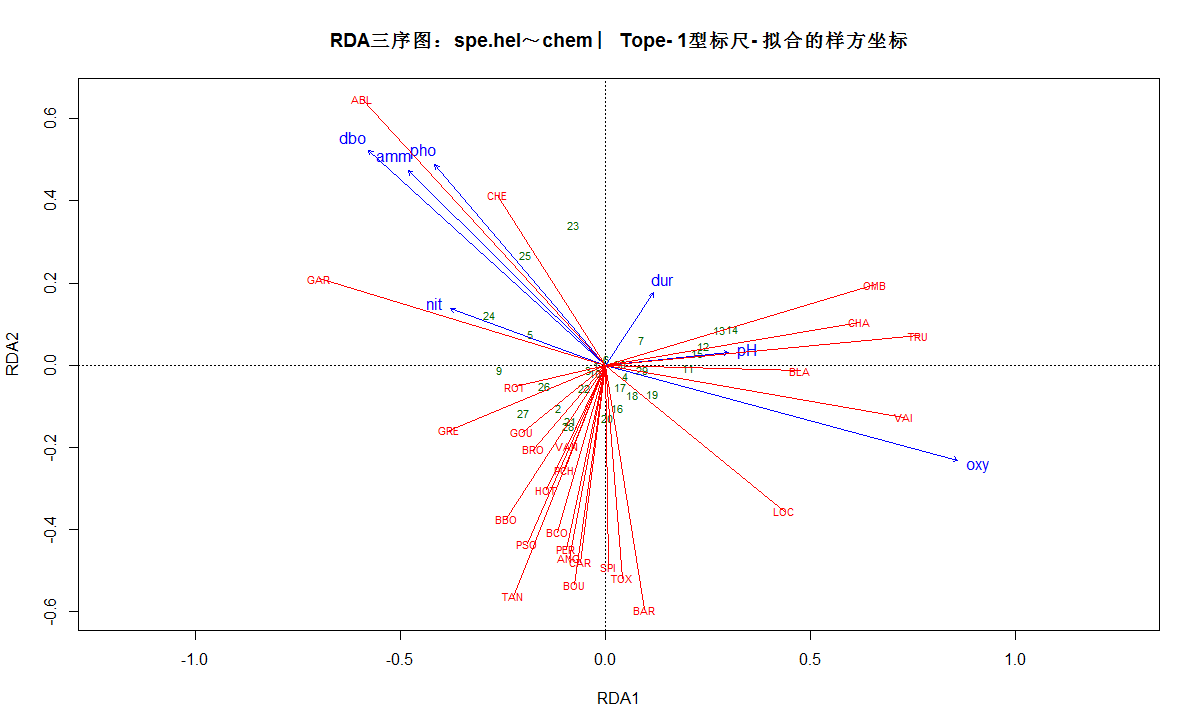
# 偏RDA三序图(使用拟合值的样方坐标)
# 1型标尺
plot(spechem.physio, scaling=1, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~chem ︳Tope- 1型标尺- 拟合的样方坐标")
spe3.sc <- scores(spechem.physio, choices=1:2, scaling=1, display="sp")
arrows(0, 0, spe3.sc[, 1], spe3.sc[, 2], length=0, lty=1, col="red")
# 2型标尺
plot(spechem.physio, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~chem ︳Tope- 2型标尺- 拟合的样方坐标")
spe4.sc <- scores(spechem.physio, choices=1:2, display="sp")
arrows(0, 0, spe4.sc[,1], spe4.sc[,2], length=0, lty=1, col="red")
解释变量向前选择
每个变量的共线性程度可以用变量的方差膨胀因子(variance inflation factor,VIF)度量,VIF是衡量一个变量的回归系数的方差由共线性引起的膨胀比例。如果VIF值超过20,表示共线性很严重。实际上,VIF超过10则可能会有共线性问题,需要处理。
# 两个RDA结果的变量方差膨胀因子(VIF)
# ********************************************
# 本章第一个RDA结果:包括所有环境因子变量
vif.cca(spe.rda)
alt penmoderate pensteep penvery_steep deb pH dur pho nit
20.397021 2.085921 2.987679 4.594983 6.684187 1.363575 3.049937 30.614913 18.953092
amm oxy dbo
40.114746 6.854703 17.980889
vif.cca(spechem.physio) # 偏RDA
alt pen deb pH dur pho nit amm oxy dbo
16.188416 1.873974 6.711460 1.205235 3.268620 25.363359 16.080319 30.685907 6.904214 17.782727
# 使用双终止准则(Blanchet等,2008a)前向选择解释变量
# 1.包含所有解释变量的RDA全模型
spe.rda.all <- rda(spe.hel ~ ., data=env)
Call: rda(formula = spe.hel ~ alt + pen + deb + pH + dur + pho + nit + amm + oxy + dbo, data = env)
Inertia Proportion Rank
Total 0.5025 1.0000
Constrained 0.3689 0.7341 10
Unconstrained 0.1336 0.2659 18
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3 RDA4 RDA5 RDA6 RDA7 RDA8 RDA9 RDA10
0.22803 0.05442 0.03382 0.03008 0.00749 0.00566 0.00443 0.00281 0.00138 0.00079
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
0.04642 0.02072 0.01746 0.01325 0.00974 0.00588 0.00512 0.00400
(Showed only 8 of all 18 unconstrained eigenvalues)
# 2.全模型校正R2
(R2a.all <- RsquareAdj(spe.rda.all)$adj.r.squared)
[1] 0.5864353
# 3.使用packfors 包内forward.sel()函数选择变量
# library(packfor) #如果尚未载入packfors包,需要运行这一步
forward.sel(spe.hel, env, adjR2thresh=R2a.all)
Testing variable 1
Testing variable 2
Testing variable 3
Testing variable 4
Procedure stopped (adjR2thresh criteria) adjR2cum = 0.594764 with 4 variables (superior to 0.586435)
variables order R2 R2Cum AdjR2Cum F pval
1 alt 1 0.32806549 0.3280655 0.3031790 13.182488 0.001
2 oxy 9 0.16402853 0.4920940 0.4530243 8.396715 0.001
3 dbo 10 0.09733024 0.5894243 0.5401552 5.926448 0.001
4 pen 2 0.06323025 0.6526545 0.5947636 4.368924 0.007
#注意,正如这个函数的提示信息所示,选择最后一个变量其实违背了
#adjR2thresh终止准则,所以pen变量最终不应该在被选变量列表中。
# 使用vegan包内ordistep()函数前向选择解释变量。该函数可以对因子变量进# 行选择,也可以运行解释变量的逐步选择和后向选择。
step.forward <- ordistep(rda(spe.hel ~ 1, data=env), scope = formula(spe.rda.all ),
direction="forward", pstep = 1000)
Start: spe.hel ~ 1
Df AIC F Pr(>F)
+ alt 1 -28.504 13.1825 0.005 **
+ oxy 1 -27.420 11.7086 0.005 **
+ deb 1 -26.872 10.9840 0.005 **
+ nit 1 -26.716 10.7795 0.005 **
+ dbo 1 -23.172 6.4340 0.005 **
+ dur 1 -22.499 5.6673 0.005 **
+ pho 1 -22.189 5.3200 0.005 **
+ amm 1 -22.047 5.1620 0.005 **
+ pen 1 -20.155 3.1305 0.005 **
+ pH 1 -17.489 0.4839 0.815
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Step: spe.hel ~ alt
Df AIC F Pr(>F)
+ oxy 1 -34.620 8.3967 0.005 **
+ dbo 1 -32.103 5.5373 0.005 **
+ amm 1 -30.777 4.1281 0.010 **
+ pho 1 -30.560 3.9032 0.010 **
+ nit 1 -29.451 2.7810 0.035 *
+ pen 1 -29.049 2.3847 0.040 *
+ deb 1 -27.972 1.3504 0.170
+ dur 1 -27.954 1.3332 0.290
+ pH 1 -27.426 0.8403 0.515
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Step: spe.hel ~ alt + oxy
Df AIC F Pr(>F)
+ dbo 1 -38.789 5.9264 0.005 **
+ pho 1 -37.052 4.1280 0.010 **
+ amm 1 -36.527 3.6055 0.015 *
+ pen 1 -36.399 3.4797 0.015 *
+ dur 1 -34.740 1.8964 0.095 .
+ deb 1 -34.388 1.5714 0.125
+ nit 1 -33.474 0.7474 0.620
+ pH 1 -33.035 0.3605 0.950
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Step: spe.hel ~ alt + oxy + dbo
Df AIC F Pr(>F)
+ pen 1 -41.639 4.3689 0.015 *
+ dur 1 -39.394 2.2555 0.025 *
+ deb 1 -38.436 1.4019 0.190
+ pho 1 -37.789 0.8420 0.455
+ nit 1 -37.577 0.6611 0.655
+ amm 1 -37.583 0.6656 0.700
+ pH 1 -37.316 0.4399 0.910
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Step: spe.hel ~ alt + oxy + dbo + pen
Df AIC F Pr(>F)
+ dur 1 -41.502 1.5255 0.130
+ deb 1 -41.058 1.1528 0.380
+ pho 1 -40.822 0.9570 0.470
+ amm 1 -40.587 0.7641 0.630
+ nit 1 -40.560 0.7418 0.635
+ pH 1 -40.375 0.5912 0.760
> RsquareAdj(rda(spe.hel ~ alt, data=env))$adj.r.squared
[1] 0.303179
> RsquareAdj(rda(spe.hel ~ alt+oxy, data=env))$adj.r.squared
[1] 0.4530243
> RsquareAdj(rda(spe.hel ~ alt+oxy+dbo, data=env))$adj.r.squared
[1] 0.5401552
> RsquareAdj(rda(spe.hel ~ alt+oxy+dbo+pen, data=env))$adj.r.squared
[1] 0.5947636
> #简约的RDA分析
> # **************
> spe.rda.pars <- rda(spe.hel ~ alt + oxy + dbo, data=env)
> spe.rda.pars
Call: rda(formula = spe.hel ~ alt + oxy + dbo, data = env)
Inertia Proportion Rank
Total 0.5025 1.0000
Constrained 0.2962 0.5894 3
Unconstrained 0.2063 0.4106 25
Inertia is variance
Eigenvalues for constrained axes:
RDA1 RDA2 RDA3
0.21802 0.05088 0.02729
Eigenvalues for unconstrained axes:
PC1 PC2 PC3 PC4 PC5 PC6 PC7 PC8
0.05835 0.04357 0.02568 0.01740 0.01451 0.01230 0.00787 0.00646
(Showed only 8 of all 25 unconstrained eigenvalues)
> anova.cca(spe.rda.pars, step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ alt + oxy + dbo, data = env)
Df Variance F Pr(>F)
Model 3 0.29619 11.963 0.001 ***
Residual 25 0.20632
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> anova.cca(spe.rda.pars, step=1000, by="axis")
Permutation test for rda under reduced model
Forward tests for axes
Permutation: free
Number of permutations: 999
Model: rda(formula = spe.hel ~ alt + oxy + dbo, data = env)
Df Variance F Pr(>F)
RDA1 1 0.218022 26.4181 0.001 ***
RDA2 1 0.050879 6.1651 0.001 ***
RDA3 1 0.027291 3.3069 0.002 **
Residual 25 0.206319
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> vif.cca(spe.rda.pars)
alt oxy dbo
1.223386 3.544201 3.402515
> (R2a.pars <- RsquareAdj(spe.rda.pars)$adj.r.squared)
[1] 0.5401552
# 1型标尺
plot(spe.rda.pars, scaling=1, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~alt+oxy+dbo - 1型标尺 - 拟合的样方坐标")
spe4.sc = scores(spe.rda.pars, choices=1:2, scaling=1, display="sp")
arrows(0, 0, spe4.sc[, 1], spe4.sc[, 2], length=0, lty=1, col="red")
# 2型标尺
plot(spe.rda.pars, display=c("sp", "lc", "cn"),
main="RDA三序图:spe.hel~alt+oxy+dbo - 2型标尺 - 拟合的样方坐标")
spe5.sc = scores(spe.rda.pars, choices=1:2, display="sp")
arrows(0, 0, spe5.sc[,1], spe5.sc[,2], length=0, lty=1, col="red")
#如果第三典范轴也显著,可以选择绘制轴1和轴3、轴2和轴3的三序图。
变差分解
# 变差分解说明图
par(mfrow=c(1,3))
showvarparts(2) # 两组解释变量
showvarparts(3) #三组解释变量
showvarparts(4) #四组解释变量
# 1.带所有环境变量的变差分解
spe.part.all <- varpart(spe.hel, envchem, envtopo)
spe.part.all
Partition of variance in RDA
Call: varpart(Y = spe.hel, X = envchem, envtopo)
Explanatory tables:
X1: envchem
X2: envtopo
No. of explanatory tables: 2
Total variation (SS): 14.07
Variance: 0.50251
No. of observations: 29
Partition table:
Df R.squared Adj.R.squared Testable
[a+b] = X1 7 0.60579 0.47439 TRUE
[b+c] = X2 3 0.41526 0.34509 TRUE
[a+b+c] = X1+X2 10 0.73414 0.58644 TRUE
Individual fractions
[a] = X1|X2 7 0.24135 TRUE
[b] 0 0.23304 FALSE
[c] = X2|X1 3 0.11205 TRUE
[d] = Residuals 0.41356 FALSE
---
Use function ‘rda’ to test significance of fractions of interest
plot(spe.part.all, digits=2)
#这些图内校正R2是正确的数字,但是韦恩图圆圈大小相同,未与R2的大小成比例。
> # 2.分别对两组环境变量进行前向选择
> spe.chem <- rda(spe.hel, envchem)
> R2a.all.chem <- RsquareAdj(spe.chem)$adj.r.squared
> forward.sel(spe.hel, envchem, adjR2thresh=R2a.all.chem, nperm=9999)
Testing variable 1
Testing variable 2
Testing variable 3
Testing variable 4
Procedure stopped (adjR2thresh criteria) adjR2cum = 0.487961 with 4 variables (superior to 0.474388)
variables order R2 R2Cum AdjR2Cum F pval
1 oxy 6 0.30247973 0.3024797 0.2766456 11.708553 0.0001
2 dbo 7 0.09015052 0.3926303 0.3459095 3.859122 0.0043
3 nit 4 0.11522115 0.5078514 0.4487936 5.852965 0.0005
4 amm 5 0.05325801 0.5611094 0.4879610 2.912325 0.0083
> spe.topo <- rda(spe.hel, envtopo)
> R2a.all.topo <- RsquareAdj(spe.topo)$adj.r.squared
> forward.sel(spe.hel, envtopo, adjR2thresh=R2a.all.topo, nperm=9999)
Testing variable 1
Testing variable 2
Testing variable 3
Procedure stopped (alpha criteria): pvalue for variable 3 is 0.228900 (superior to 0.050000)
variables order R2 R2Cum AdjR2Cum F pval
1 alt 1 0.32806549 0.3280655 0.3031790 13.182488 0.0001
2 pen 2 0.05645105 0.3845165 0.3371717 2.384674 0.0469
> # 解释变量简约组合(基于变量选择的结果)
> names(env)
[1] "alt" "pen" "deb" "pH" "dur" "pho" "nit" "amm" "oxy" "dbo"
> envchem.pars <- envchem[, c(4,6,7)]
> envtopo.pars <- envtopo[, c(1,2)]
> # 变差分解
> (spe.part <- varpart(spe.hel, envchem.pars, envtopo.pars))
Partition of variance in RDA
Call: varpart(Y = spe.hel, X = envchem.pars, envtopo.pars)
Explanatory tables:
X1: envchem.pars
X2: envtopo.pars
No. of explanatory tables: 2
Total variation (SS): 14.07
Variance: 0.50251
No. of observations: 29
Partition table:
Df R.squared Adj.R.squared Testable
[a+b] = X1 3 0.50785 0.44879 TRUE
[b+c] = X2 2 0.38452 0.33717 TRUE
[a+b+c] = X1+X2 5 0.66351 0.59036 TRUE
Individual fractions
[a] = X1|X2 3 0.25318 TRUE
[b] 0 0.19561 FALSE
[c] = X2|X1 2 0.14156 TRUE
[d] = Residuals 0.40964 FALSE
---
Use function ‘rda’ to test significance of fractions of interest
> plot(spe.part, digits=2)
> # 所有可测部分的置换检验
> # [a+b]部分的检验
> anova.cca(rda(spe.hel, envchem.pars), step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.hel, Y = envchem.pars)
Df Variance F Pr(>F)
Model 3 0.25520 8.5992 0.001 ***
Residual 25 0.24731
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> # [b+c]部分的检验
> anova.cca(rda(spe.hel, envtopo.pars), step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.hel, Y = envtopo.pars)
Df Variance F Pr(>F)
Model 2 0.19322 8.1216 0.001 ***
Residual 26 0.30929
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> # [a+b+c]部分的检验
> env.pars <- cbind(envchem.pars, envtopo.pars)
> anova.cca(rda(spe.hel, env.pars), step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.hel, Y = env.pars)
Df Variance F Pr(>F)
Model 5 0.33342 9.0704 0.001 ***
Residual 23 0.16909
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> # [a]部分的检验
> anova.cca(rda(spe.hel, envchem.pars, envtopo.pars), step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.hel, Y = envchem.pars, Z = envtopo.pars)
Df Variance F Pr(>F)
Model 3 0.14020 6.3565 0.001 ***
Residual 23 0.16909
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> # [c]部分的检验
> anova.cca(rda(spe.hel, envtopo.pars, envchem.pars), step=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.hel, Y = envtopo.pars, Z = envchem.pars)
Df Variance F Pr(>F)
Model 2 0.078219 5.3197 0.001 ***
Residual 23 0.169091
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> #各个部分置换检验有不显著的吗?
> # 3.无变量nit(硝酸盐浓度)的变差分解
> envchem.pars2 <- envchem[, c(6,7)]
> (spe.part2 <- varpart(spe.hel, envchem.pars2, envtopo.pars))
Partition of variance in RDA
Call: varpart(Y = spe.hel, X = envchem.pars2, envtopo.pars)
Explanatory tables:
X1: envchem.pars2
X2: envtopo.pars
No. of explanatory tables: 2
Total variation (SS): 14.07
Variance: 0.50251
No. of observations: 29
Partition table:
Df R.squared Adj.R.squared Testable
[a+b] = X1 2 0.39263 0.34591 TRUE
[b+c] = X2 2 0.38452 0.33717 TRUE
[a+b+c] = X1+X2 4 0.65265 0.59476 TRUE
Individual fractions
[a] = X1|X2 2 0.25759 TRUE
[b] 0 0.08832 FALSE
[c] = X2|X1 2 0.24885 TRUE
[d] = Residuals 0.40524 FALSE
---
Use function ‘rda’ to test significance of fractions of interest
> plot(spe.part2, digits=2)
RDA 作为多元方差分析(MANOVA)的工具
# 基于RDA的双因素MANOVA
# **************************
# 生成代表海拔的因子变量(3个水平,每个水平含9个样方)
alt.fac <- gl(3, 9)
# 生成近似模拟pH值的因子变量
pH.fac <- as.factor(c(1, 2, 3, 2, 3, 1, 3, 2, 1, 2, 1, 3, 3, 2, 1, 1, 2, 3,
2, 1, 2, 3, 2, 1, 1, 3, 3))
# 两个因子是否平衡?
table(alt.fac, pH.fac)
table(alt.fac, pH.fac)
# 用Helmert对照法编码因子和它们的交互作用项
alt.pH.helm <- model.matrix(~ alt.fac * pH.fac,
contrasts=list(alt.fac="contr.helmert", pH.fac="contr.helmert"))
alt.pH.helm
(Intercept) alt.fac1 alt.fac2 pH.fac1 pH.fac2 alt.fac1:pH.fac1 alt.fac2:pH.fac1 alt.fac1:pH.fac2 alt.fac2:pH.fac2
1 1 -1 -1 -1 -1 1 1 1 1
2 1 -1 -1 1 -1 -1 -1 1 1
3 1 -1 -1 0 2 0 0 -2 -2
4 1 -1 -1 1 -1 -1 -1 1 1
5 1 -1 -1 0 2 0 0 -2 -2
6 1 -1 -1 -1 -1 1 1 1 1
7 1 -1 -1 0 2 0 0 -2 -2
8 1 -1 -1 1 -1 -1 -1 1 1
9 1 -1 -1 -1 -1 1 1 1 1
10 1 1 -1 1 -1 1 -1 -1 1
11 1 1 -1 -1 -1 -1 1 -1 1
12 1 1 -1 0 2 0 0 2 -2
13 1 1 -1 0 2 0 0 2 -2
14 1 1 -1 1 -1 1 -1 -1 1
15 1 1 -1 -1 -1 -1 1 -1 1
16 1 1 -1 -1 -1 -1 1 -1 1
17 1 1 -1 1 -1 1 -1 -1 1
18 1 1 -1 0 2 0 0 2 -2
19 1 0 2 1 -1 0 2 0 -2
20 1 0 2 -1 -1 0 -2 0 -2
21 1 0 2 1 -1 0 2 0 -2
22 1 0 2 0 2 0 0 0 4
23 1 0 2 1 -1 0 2 0 -2
24 1 0 2 -1 -1 0 -2 0 -2
25 1 0 2 -1 -1 0 -2 0 -2
26 1 0 2 0 2 0 0 0 4
27 1 0 2 0 2 0 0 0 4
attr(,"assign")
[1] 0 1 1 2 2 3 3 3 3
attr(,"contrasts")
attr(,"contrasts")$alt.fac
[1] "contr.helmert"
attr(,"contrasts")$pH.fac
[1] "contr.helmert"
#检查当前对照法产生的表格,哪一列代表海拔因子、pH值和交互作用项?
# 检查Helmert对照表属性1:每个变量的和为1
apply(alt.pH.helm[, 2:9], 2, sum)
# 检查Helmert对照表属性2:变量之间不相关
cor(alt.pH.helm[, 2:9])
alt.fac1 alt.fac2 pH.fac1 pH.fac2 alt.fac1:pH.fac1 alt.fac2:pH.fac1 alt.fac1:pH.fac2 alt.fac2:pH.fac2
alt.fac1 1 0 0 0 0 0 0 0
alt.fac2 0 1 0 0 0 0 0 0
pH.fac1 0 0 1 0 0 0 0 0
pH.fac2 0 0 0 1 0 0 0 0
alt.fac1:pH.fac1 0 0 0 0 1 0 0 0
alt.fac2:pH.fac1 0 0 0 0 0 1 0 0
alt.fac1:pH.fac2 0 0 0 0 0 0 1 0
alt.fac2:pH.fac2 0 0 0 0 0 0 0 1
# 使用函数betadisper()(vegan包)(Marti Anderson检验)验证组内协方差矩阵# 的齐性
spe.hel.d1 <- dist(spe.hel[1:27,])
# 海拔因子
(spe.hel.alt.MHV <- betadisper(spe.hel.d1, alt.fac))
Homogeneity of multivariate dispersions
Call: betadisper(d = spe.hel.d1, group = alt.fac)
No. of Positive Eigenvalues: 26
No. of Negative Eigenvalues: 0
Average distance to median:
1 2 3
0.5208 0.5175 0.3881
Eigenvalues for PCoA axes:
PCoA1 PCoA2 PCoA3 PCoA4 PCoA5 PCoA6 PCoA7 PCoA8
6.5329 1.7407 1.2269 1.0591 0.6117 0.4683 0.3987 0.3207
anova(spe.hel.alt.MHV)
Analysis of Variance Table
Response: Distances
Df Sum Sq Mean Sq F value Pr(>F)
Groups 2 0.1032 0.051602 0.8763 0.4292
Residuals 24 1.4133 0.058889
permutest(spe.hel.alt.MHV) # 置换检验
Permutation test for homogeneity of multivariate dispersions
Permutation: free
Number of permutations: 999
Response: Distances
Df Sum Sq Mean Sq F N.Perm Pr(>F)
Groups 2 0.1032 0.051602 0.8763 999 0.439
Residuals 24 1.4133 0.058889
> # pH值因子
> (spe.hel.pH.MHV <- betadisper(spe.hel.d1, pH.fac))
Homogeneity of multivariate dispersions
Call: betadisper(d = spe.hel.d1, group = pH.fac)
No. of Positive Eigenvalues: 26
No. of Negative Eigenvalues: 0
Average distance to median:
1 2 3
0.6658 0.6716 0.7019
Eigenvalues for PCoA axes:
PCoA1 PCoA2 PCoA3 PCoA4 PCoA5 PCoA6 PCoA7 PCoA8
6.5329 1.7407 1.2269 1.0591 0.6117 0.4683 0.3987 0.3207
> anova(spe.hel.pH.MHV)
Analysis of Variance Table
Response: Distances
Df Sum Sq Mean Sq F value Pr(>F)
Groups 2 0.00676 0.0033802 0.1587 0.8542
Residuals 24 0.51124 0.0213018
> permutest(spe.hel.pH.MHV) #置换检验
Permutation test for homogeneity of multivariate dispersions
Permutation: free
Number of permutations: 999
Response: Distances
Df Sum Sq Mean Sq F N.Perm Pr(>F)
Groups 2 0.00676 0.0033802 0.1587 999 0.855
Residuals 24 0.51124 0.0213018
> #组内协方差齐性,可以继续分析。
# 首先检验交互作用项。海拔因子和pH因子构成协变量矩阵
interaction.rda <- rda(spe.hel[1:27, ], alt.pH.helm[, 6:9], alt.pH.helm[, 2:5])
anova(interaction.rda, step=1000, perm.max=1000)
#交互作用是否显著?显著的交互作用表示一个因子的影响依赖另一个因子
#的水平,这将妨害主因子变量的分析。
# 检验海拔因子的效应,此时pH值因子和交互作用项作为协变量矩阵
factor.alt.rda <- rda(spe.hel[1:27, ], alt.pH.helm[, 2:3], alt.pH.helm[, 4:9])
anova(factor.alt.rda, step=1000, perm.max=1000, strata=pH.fac)
#海拔因子影响是否显著?
#检验pH值因子的效应,此时海拔值因子和交互作用项作为协变量矩阵
factor.pH.rda <- rda(spe.hel[1:27, ], alt.pH.helm[, 4:5],
alt.pH.helm[, c(2:3, 6:9)])
anova(factor.pH.rda, step=1000, perm.max=1000, strata=alt.fac)
#pH值影响是否显著?
# RDA和显著影响的海拔因子三序图
alt.rda.out <- rda(spe.hel[1:27,]~., as.data.frame(alt.fac))
plot(alt.rda.out, scaling=1, display=c("sp", "wa", "cn"),
main="Multivariate ANOVA, factor altitude - scaling 1 - wa scores")
spe.manova.sc <- scores(alt.rda.out, choices=1:2, scaling=1, display="sp")
arrows(0, 0, spe.manova.sc[, 1], spe.manova.sc[, 2], length=0, col="red")
基于距离的RDA分析
> # 基于距离的RDA分析(db-RDA)
> # ****************************
> # 1.分步计算
> spe.bray <- vegdist(spe[1:27, ], "bray")
> spe.pcoa <- cmdscale(spe.bray, k=nrow(spe[1:27, ])-1, eig=TRUE, add=TRUE)
> spe.scores <- spe.pcoa$points
> # 交互作用的检验。从协变量矩阵获得Helmert对照编码海拔因子和pH值因子
> interact.dbrda <- rda(spe.scores[1:27, ], alt.pH.helm[, 6:9], alt.pH.helm[, 2:5])
> anova(interact.dbrda, step=1000, perm.max=1000)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(X = spe.scores[1:27, ], Y = alt.pH.helm[, 6:9], Z = alt.pH.helm[, 2:5])
Df Variance F Pr(>F)
Model 4 0.021857 0.441 1
Residual 18 0.223016
> #交互作用是否显著?如果没有,可以继续检验主因子的效应(此处未显示)
> # 2.直接使用vegan包内capscale()函数运行。只能以模型界面运行。响应变量
> #可以是原始数据矩阵。
> interact.dbrda2 <- capscale(spe[1:27,] ~ alt.fac*pH.fac + Condition(alt.fac+pH.fac), distance="bray", add=TRUE)
> anova(interact.dbrda2, step=1000, perm.max=1000)
Permutation test for capscale under reduced model
Permutation: free
Number of permutations: 999
Model: capscale(formula = spe[1:27, ] ~ alt.fac * pH.fac + Condition(alt.fac + pH.fac), distance = "bray", add = TRUE)
Df SumOfSqs F Pr(>F)
Model 4 0.4667 0.4811 1
Residual 18 4.3658
> # 或者响应变量可以是相异矩阵。
> interact.dbrda3 <- capscale(spe.bray ~ alt.fac*pH.fac + Condition(alt.fac+pH.fac), add=TRUE)
> anova(interact.dbrda3, step=1000, perm.max=1000)
Permutation test for capscale under reduced model
Permutation: free
Number of permutations: 999
Model: capscale(formula = spe.bray ~ alt.fac * pH.fac + Condition(alt.fac + pH.fac), add = TRUE)
Df SumOfSqs F Pr(>F)
Model 4 0.4667 0.4811 1
Residual 18 4.3658
非线性关系的RDA分析
> # 二阶解释变量的RDA
> # *******************
> # 生成das和das正交二阶项(由poly()函数获得)矩阵
> das.df <- poly(das, 2)
> colnames(das.df) <- c("das", "das2")
> # 验证两个变量是否显著
> forward.sel(spe, das.df)
Testing variable 1
Testing variable 2
variables order R2 R2Cum AdjR2Cum F pval
1 das 1 0.44777219 0.4477722 0.4273193 21.892865 0.001
2 das2 2 0.07870749 0.5264797 0.4900550 4.321662 0.005
> # RDA和置换检验
> spe.das.rda <- rda(spe ~ ., as.data.frame(das.df))
> anova(spe.das.rda)
Permutation test for rda under reduced model
Permutation: free
Number of permutations: 999
Model: rda(formula = spe ~ das + das2, data = as.data.frame(das.df))
Df Variance F Pr(>F)
Model 2 36.304 14.454 0.001 ***
Residual 26 32.652
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
> # 三序图(拟合的样方坐标,2型标尺)
> plot(spe.das.rda, scaling=2, display=c("sp", "lc", "cn"),
+ main="RDA三序图:spe~das + das2 - 2型标尺 - 拟合的样方坐标")
> spe6.sc <- scores(spe.das.rda, choices=1:2, scaling=2, display="sp")
> arrows(0, 0, spe6.sc[, 1], spe6.sc[, 2], length=0, lty=1, col="red")
# 4种鱼类的分布地图
# ******************
par(mfrow=c(2, 2))
plot(spa$x, spa$y, asp=1, col="brown", cex=spe$TRU,
xlab="x (km)", ylab="y (km)", main="褐鳟")
lines(spa$x, spa$y, col="light blue")
plot(spa$x, spa$y, asp=1, col="brown", cex=spe$OMB,
xlab="x (km)", ylab="y (km)", main="鳟鱼")
lines(spa$x, spa$y, col="light blue")
plot(spa$x, spa$y, asp=1, col="brown", cex=spe$ABL,
xlab="x (km)", ylab="y (km)", main="欧鮊鱼")
lines(spa$x, spa$y, col="light blue")
plot(spa$x, spa$y, asp=1, col="brown", cex=spe$TAN,
xlab="x (km)", ylab="y (km)", main="鲤鱼")
lines(spa$x, spa$y, col="light blue")
自写代码角
为了能够正确自写RDA分析代码,有必要参考Legendre和Legendre
(1998)第11.1节相关内容。下面是计算步骤(基于协方差矩阵的RDA)
1.计算中心化的物种数据矩阵与标准化解释变量矩阵的多元线性回归,获得拟合值矩阵;
2.计算拟合值矩阵的PCA;
3.计算两类样方坐标;
4.结果输出。
下面代码解释部分使用的公式编码与Legendre和Legendre(1998)一致。
下面的代码集中在RDA约束部分,目的是使读者对RDA数学过程感兴趣,而不是最优化程序。
myRDA <- function(Y,X) {
# 1.数据的准备
# *************
Y.mat <- as.matrix(Y)
Yc <- scale(Y.mat, scale=FALSE)
X.mat <- as.matrix(X)
Xcr <- scale(X.mat)
# 2.多元线性回归的计算
# *********************
# 回归系数矩阵 (eq. 11.4)
B <- solve(t(Xcr) %*% Xcr) %*% t(Xcr) %*% Yc
# 拟合值矩阵(eq. 11.5)
Yhat <- Xcr %*% B
# 残差矩阵
Yres <- Yc - Yhat
#维度
n <- nrow(Y)
p <- ncol(Y)
m <- ncol(X)
# 3. 拟合值PCA分析
# ******************
# 协方差矩阵 (eq. 11.7)
S <- cov(Yhat)
# 特征根分解
eigenS <- eigen(S)
# 多少个典范轴?
kc <- length(which(eigenS$values > 0.00000001))
# 典范轴特征根
ev <- eigenS$values[1:kc]
# 矩阵Yc(中心化)的总方差(惯量)
trace = sum(diag(cov(Yc)))
# 正交特征向量(响应变量的贡献,1型标尺)
U <- eigenS$vectors[,1:kc]
row.names(U) <- colnames(Y)
# 样方坐标(vegan包内'wa' 坐标,1型标尺eq. 11.12)
F <- Yc %*% U
row.names(F) <- row.names(Y)
# 样方约束(vegan包内'lc' 坐标,2型标尺eq. 11.13)
Z <- Yhat %*% U
row.names(Z) <- row.names(Y)
# 典范系数 (eq. 11.14)
CC <- B %*% U
row.names(CC) <- colnames(X)
# 解释变量
# 物种-环境相关
corXZ <- cor(X,Z)
# 权重矩阵的诊断
D <- diag(sqrt(ev/trace))
# 解释变量双序图坐标
coordX <- corXZ %*% D #1型标尺
coordX2 <- corXZ #2型标尺
row.names(coordX) <- colnames(X)
row.names(coordX2) <- colnames(X)
# 相对特征根平方根转化(为2型标尺)
U2 <- U %*% diag(sqrt(ev))
row.names(U2) <- colnames(Y)
F2 <- F %*% diag(1/sqrt(ev))
row.names(F2) <- row.names(Y)
Z2 <- Z %*% diag(1/sqrt(ev))
row.names(Z2) <- row.names(Y)
# 未校正R2
R2 <- sum(ev/trace)
# 校正R2
R2adj <- 1-((n-1)/(n-m-1))*(1-R2)
# 4.残差的PCA
# *******************
# 与第5章相同,写自己的代码,可以从这里开始...
# eigenSres <- eigen(cov(Yres))
# evr <- eigenSres$values
# 5.输出Output
result <- list(trace, R2, R2adj,ev,CC,U,F,Z,coordX, U2,F2,Z2,coordX2)
names(result) <- c("Total_variance", "R2", "R2a","Can_ev", "Can_coeff",
"Species_sc1", "wa_sc1", "lc_sc1", "Biplot_sc1", "Species_sc2",
"wa_sc2", "lc_sc2", "Biplot_sc2")
result
}
#将此函数应用到Doubs鱼类数据和环境数据的RDA分析
doubs.myRDA <- myRDA(spe.hel,env)
summary(doubs.myRDA)
Length Class Mode
Total_variance 1 -none- numeric
R2 1 -none- numeric
R2a 1 -none- numeric
Can_ev 10 -none- numeric
Can_coeff 100 -none- numeric
Species_sc1 270 -none- numeric
wa_sc1 290 -none- numeric
lc_sc1 290 -none- numeric
Biplot_sc1 100 -none- numeric
Species_sc2 270 -none- numeric
wa_sc2 290 -none- numeric
lc_sc2 290 -none- numeric
Biplot_sc2 100 -none- numeric
置换检验(R语言实现)