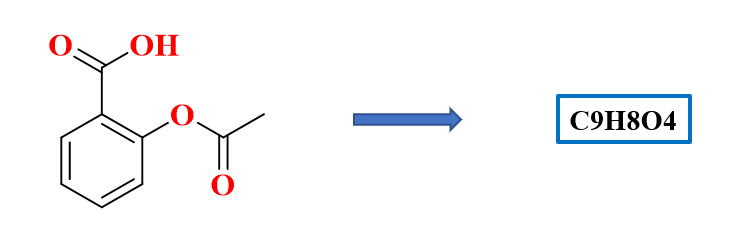
RDKit | 基于化合物结构式图像估算分子式
1
简介
当通过深度学习输入有机物质中结构式的二维图像时,需要解决寻找分子式的问题。这是一个回归问题,需要计算结构式图像中包含的碳、氢、氧和氮等原子数。
2
环境
系统 :Win10
工具:RDKit、OpenCV、Keras、TensorFlow
3
实验步骤
训练数据300,000种化合物的SMILES字符串(足够的训练数据)。
输入结构的二维图像可以提供足够的信息来理解分子结构。
使用RDKit将SMILES字符串转换为结构式图像,并进行学习以计算图像中的原子数。
4
数据预处理
提取获得类似如下数据
5
示例代码
导入库
import keras
from keras.layers import Input, Concatenate, GlobalAveragePooling2D
from keras.models import Model
from keras.layers.core import Dense, Dropout, Activation, Flatten
from keras.callbacks import LearningRateScheduler, ModelCheckpoint
from keras.preprocessing.image import img_to_array, array_to_img
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import os.path
import re
import tensorflow as tf
from keras import backend as K
import cv2
from rdkit import Chem
from rdkit.Chem import Draw, rdDepictor, rdMolDescriptors
import warnings
warnings.filterwarnings("ignore")
载入数据
#Load SMILES
df = pd.read_csv('data.csv')
SMILES = df['CAN_SMILES'].values
SMILES转numpy
# convert to array
SMILES = np.asarray(SMILES)
SMILES_train, SMILES_test = train_test_split(SMILES, test_size=0.30, random_state=110)
print(SMILES_train.shape, SMILES_test.shape)
(210000,) (90000,)
定义DataGenerator数据生成函数
#Release memory for read data
del df, SMILES
class DataGenerator():
def __init__(self, X_input):
self.X_input = X_input
self.atom = ['C', 'H', 'N', 'O', 'S', 'P', 'Si', 'Na', 'F', 'Cl', 'Br', 'I']
self.reset()
def reset(self):
self.X = []
self.Y = []
def cal_img(self, mol):
# Generate structural formula image
img = Chem.Draw.MolToImage(mol, size=(300, 300))
return img_to_array(img)[:,:,:3]
def chem_count(self, input):
atom = ['C', 'H', 'N', 'O', 'S', 'P', 'Si', 'Na', 'F', 'Cl', 'Br', 'I']
count = {'C': 0, 'H': 0 ,'N': 0, 'O': 0, 'S': 0, 'P': 0, 'Si': 0, 'Na': 0, 'F': 0, 'Cl': 0, 'Br': 0, 'I': 0}
input = input + 'Z' # Add English capital letters at the end
for i in range(len(atom)):
if re.search(atom[i] + '\d{1,3}[A-Z]', input): #Atom + number (1 to 3 digits) + uppercase English
iterator = re.finditer(atom[i] + '\d{1,3}[A-Z]', input)
for match in iterator:
count[atom[i]] = int(match.group()[len(atom[i]):-1])
elif re.search(atom[i] + '[A-Z]',input): #When containing atoms + uppercase English
count[atom[i]] = 1
return count
def make_structural_image(self, batch_size=10):
while True:
for i in range(len(self.X_input)):
# SConvert SMILES to mol
mol = Chem.MolFromSmiles(self.X_input[i], sanitize=True)
if not mol is None: # mol file is None
# Calculate 2D coordinates
rdDepictor.Compute2DCoords(mol)
# CalcMolWt
MW = Chem.rdMolDescriptors._CalcMolWt(mol)
# Calculate molecular formula from mol format
chem_Formula = Chem.rdMolDescriptors.CalcMolFormula(mol)
# Count atoms from molecular formula
count = self.chem_count(chem_Formula)
# Add X, Y
if MW < max_MW and count['C'] > 1.0: # A compound containing at least two carbon atoms and having a molar mass of 400 or less.
(self.X).append(self.cal_img(mol))
(self.Y).append([count[self.atom[i]] for i in range(len(self.atom))])
# Returns the value of X, Y
if len(self.X) == batch_size:
inputs = np.asarray(self.X)
outputs = np.asarray(self.Y)
self.reset()
yield inputs, outputs
建立CNN
使用RDKit读取SMILES数据并将其转换为(300 ,300 ,3 )大小的图像。同样,输出分子式['C','H','N','O','S','P','Si','Na','F','Cl','Br' ,'I']是12种原子的数量。
from keras.applications.xception import Xception
inputs = Input(shape=(300,300,3)) # data_size (300,300,3)
x = Xception(weights=None,input_shape=(300,300,3), include_top=False, pooling='avg')(inputs)
y = Dense(12, activation='linear')(x)
model = Model(inputs, y)
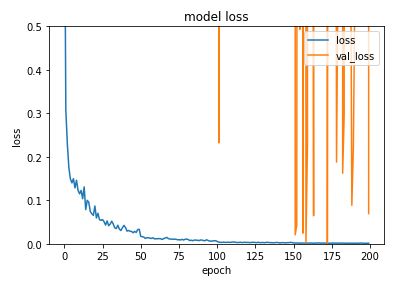
训练模型并绘制训练曲线
# Learning
history = model.fit_generator(
generator=train_datagen.make_structural_image(batch_size=batchsize),
steps_per_epoch=int(len(SMILES_train) / batchsize / 100),
epochs=epochs,
verbose=1,
validation_data=test_datagen.make_structural_image(batch_size=batchsize),
validation_steps=100,
callbacks=callbacks)
#Plot training graph
plt.figure()
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.xlabel('epoch')
plt.ylabel('loss')
plt.legend(['loss', 'val_loss'], loc='upper right')
plt.ylim([0,0.5])
plt.show()
预测测试结果输出
#Output 50 test results
atom = ['C', 'H', 'N', 'O', 'S', 'P', 'Si', 'Na', 'F', 'Cl', 'Br', 'I']
for i in range(50):
img = array_to_img(X_test[i])
real = ''
pred = ''
for j in range(len(atom)):
if round(Y_test[i,j]) > 0:
real += atom[j] + str(round(Y_test[i,j]))
if round(Y_predict_test[i,j],1) > 0.5:
pred += atom[j] + str(round(Y_predict_test[i,j],1))
img.save('test_result%04d_%s_%s.png' % (i, real, pred))
参考资料
1. http://www.rdkit.org/docs/GettingStartedInPython.html
2.https://blog.csdn.net/u012325865/article/details/82318084utm_source=blogxgwz3
3. https://blog.csdn.net/u012325865/article/details/103044562
4. https://blog.csdn.net/qq_35082030/article/details/79144374
5. https://blog.csdn.net/qq_19332527/article/details/79829087
作者&编辑
王建民
DrugAI
长按识别二维码关注我们获取最新消息!
本文为DrugAI原创编译整理,如需转载,请在公众号后台留言。