Purpose:
To validate the cell type enrichment results from the xCell, do correlation analysis to see the trend between expected each cell type proportions and real each cell type counts.
1. Data preprocessing
- transform row and column of xcell resuts
a <- read.table("xCell__xCell_0412052318.txt",header = TRUE,sep="\t")
b <- t(a)
write.table(b,file="xCell_res2",quote = FALSE,sep="\t",col.names = FALSE)
- combined xCell results with bloodCount to make new matrix
paste bloodCounts_2 xCell_res2 > cor_input
awk '{$39="";print $0}' cor_input > cor_input_2
sed 's/\t\t/\t/g' cor_input_2 > cor_input_3
2. Correlation analysis using cor function or rcorr function in Hmisc package
- Load data
- all data of cell type
setwd("F:/sdc_project")
mydata <- read.table("cor_input_3",header = TRUE,sep="\t",row.names = 1)
> mydata[1:4,1:8]
BAS BA CCMH EOS EO HB HCM HE
31201 0.4 0.02 345 2.9 0.15 162 32.9 4.93
31202 0.3 0.02 315 1.5 0.10 130 26.3 4.95
31303 0.6 0.03 327 1.9 0.09 148 27.4 5.40
31304 0.3 0.02 330 2.0 0.12 146 29.7 4.92
> dim(mydata)
[1] 915 104
- Cell type in bloodCounts realsted xCell results
part_data <- mydata[,c(1,2,4,5,8,11,16,17,18,19,20,23,24,41,57,59,76,81,85)]
> part_data[1:4,]
BAS BA EOS EO HE LE MON MOt NEU NE PTES RETIS RETAB Basophils
31201 0.4 0.02 2.9 0.15 4.93 5.16 10.3 0.53 60.2 3.11 163 1.31 0.0646 0.6411
31202 0.3 0.02 1.5 0.10 4.95 6.85 6.6 0.45 62.0 4.25 156 1.50 0.0743 0.1741
31303 0.6 0.03 1.9 0.09 5.40 4.77 6.3 0.30 59.1 2.82 201 0.96 0.0518 0.6074
31304 0.3 0.02 2.0 0.12 4.92 5.89 6.6 0.39 50.7 2.98 214 1.08 0.0531 0.0312
Eosinophils Erythrocytes Monocytes Neutrophils Platelets
31201 0.0918 0.1071 0.1476 0.0000 0.1022
31202 0.3104 0.1849 0.0276 0.1492 0.2070
31303 0.1864 0.0817 0.0000 0.0745 0.2708
31304 0.0000 0.0000 0.0000 0.0000 0.1819
> dim(part_data)
[1] 915 19
- Calculate the correlation coefficient matrix
- use cor function
# (1) use cor function
res <- cor(mydata)
res <- round(res, 2)
res[1:4,39:49]
part_res <- cor(part_data)
part_res <- round(part_res, 2)
part_res[1:4,]
- use rcorr() function
cor() function only can calculate correlation coefficient,but can't provide significant p -value, rcorr() function in Hmisc package can calculate both correlation coefficient and pvalue .
usege : rcorr(x, type =c(“pearson”,“spearman”))。
# install the package
source('http://bioconductor.org/biocLite.R')
biocLite("Hmisc")
library(Hmisc)
res2 <- rcorr(as.matrix(mydata))
res2
# part_data
part_res2 <- rcorr(as.matrix(part_data))
part_res2
# use res2$r、res2$P to get correlation coefficient and p-value
res2$r
res2$P
# part_data
part_res2$r
part_res2$P
> part_res2
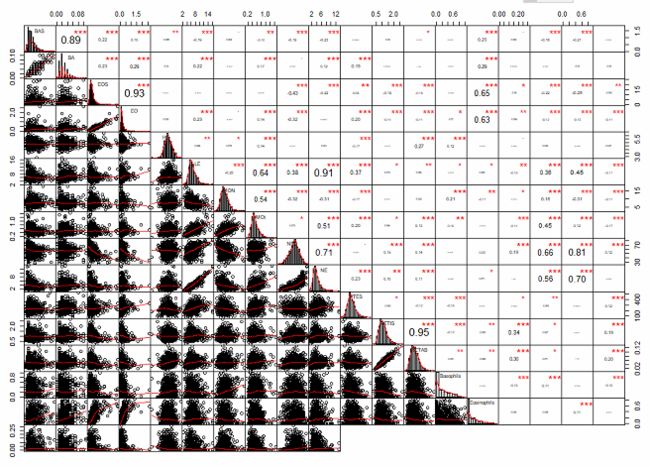
BAS BA EOS EO HE LE MON MOt NEU NE PTES RETIS
BAS 1.00 0.89 0.22 0.15 -0.09 -0.19 0.06 -0.10 -0.19 -0.23 0.01 -0.05
BA 0.89 1.00 0.23 0.29 -0.04 0.22 -0.03 0.17 -0.06 0.12 0.19 0.00
EOS 0.22 0.23 1.00 0.93 0.01 -0.03 0.01 -0.02 -0.43 -0.22 0.09 -0.19
EO 0.15 0.29 0.93 1.00 0.03 0.23 -0.06 0.14 -0.32 0.01 0.20 -0.14
HE -0.09 -0.04 0.01 0.03 1.00 0.09 0.07 0.14 -0.03 0.05 -0.17 -0.02
LE -0.19 0.22 -0.03 0.23 0.09 1.00 -0.25 0.64 0.38 0.91 0.37 0.07
MON 0.06 -0.03 0.01 -0.06 0.07 -0.25 1.00 0.54 -0.32 -0.31 -0.17 0.02
MOt -0.10 0.17 -0.02 0.14 0.14 0.64 0.54 1.00 0.07 0.51 0.20 0.09
NEU -0.19 -0.06 -0.43 -0.32 -0.03 0.38 -0.32 0.07 1.00 0.71 -0.06 0.15
NE -0.23 0.12 -0.22 0.01 0.05 0.91 -0.31 0.51 0.71 1.00 0.23 0.10
PTES 0.01 0.19 0.09 0.20 -0.17 0.37 -0.17 0.20 -0.06 0.23 1.00 -0.08
RETIS -0.05 0.00 -0.19 -0.14 -0.02 0.07 0.02 0.09 0.15 0.10 -0.08 1.00
RETAB -0.07 0.00 -0.18 -0.11 0.27 0.10 0.04 0.13 0.14 0.11 -0.12 0.95
Basophils 0.01 -0.02 -0.04 -0.07 0.12 -0.08 0.21 0.10 0.02 -0.05 -0.16 -0.12
3. Visualization
#abbr.colnames
symnum(res, abbr.colnames = FALSE)
symnum(part_res, abbr.colnames = FALSE)
biocLite("corrplot")
library(corrplot)
corrplot(res, type = "upper", tl.col = "black", tl.srt = 45)
corrplot(part_res, type = "upper", tl.col = "black", tl.srt = 45)
- Combined correlation coefficient and P-value
# Insignificant correlations are leaved blank
corrplot(res2$r, type="upper",p.mat = res2$P, sig.level = 0.01, tl.col = "black", insig = "blank")
corrplot(part_res2$r, order="hclust",type="upper",p.mat = res2$P, sig.level = 0.05, tl.col = "black",insig = "blank")
corrplot.mixed(part_res2$r, order="hclust",p.mat = res2$P, sig.level = 0.05, tl.col = "black",insig = "blank")
- Use chart.Correlation(): Draw scatter plots
Use chart.Correlation() in PerformanceAnalytics package
biocLite("PerformanceAnalytics")
library(PerformanceAnalytics)#加载包
chart.Correlation(mydata, histogram=TRUE, pch=19)
chart.Correlation(part_data, histogram=TRUE, pch=19)
- heatmap
col<- colorRampPalette(c("blue", "white", "red"))(20)
heatmap(x = res2$r, col = col, symm = TRUE)
heatmap(x=part_res2$r,col=col,symn=TRUE)