IsolationForest-02Python案例
Intro
sklearn中IsolationForest使用,包括参数说明和实际案例。
简述下算法思想: 随机选择特征,在该特征的maximum和minimum中随机选择切分值(split value)。如此递归划分,形成树。根节点到终止节点(叶子结点)的长度,等价于split的次数。对于多棵树,计算平均长度,可以反映样本异常的程度。即异常样本通常较快被划分到叶子结点,因而路径长度较小。
slearn版本:0.22
import sklearn
sklearn.__version__
'0.22'
参数介绍
Parameters
n_estimators:int, optional (default=100)
//树的棵数,paper中建议100棵,再增加模型效果提升有限//
The number of base estimators in the ensemble.
max_samples:int or float, optional (default=”auto”)
//sunsample样本大小,默认256,如果是int,则抽取样本数为该值;如果是float,按照比例计算即可//
The number of samples to draw from X to train each base estimator.
- If int, then draw max_samples samples.
- If float, then draw max_samples * X.shape[0] samples.
- If “auto”, then max_samples=min(256, n_samples).
If max_samples is larger than the number of samples provided, all samples will be used for all trees (no sampling).
contamination:‘auto’ or float, optional (default=‘auto’)
//样本中离群值的占比,默认"auto"是采用paper中的阈值,paper似乎没有提阈值,sklearn是0.5;float则自己指定,范围[0,0.5],这里float是分位数的意思,0.25就是得到所有得分,计算25%分位数 //
The amount of contamination of the data set, i.e. the proportion of outliers in the data set. Used when fitting to define the threshold on the scores of the samples.
- If ‘auto’, the threshold is determined as in the original paper.
- If float, the contamination should be in the range [0, 0.5].
max_features:int or float, optional (default=1.0)
//每棵树训练时,参与分裂的树的特征数,默认是1,代码里看好像是不用进行列抽样,但是bagging那里的代码没怎么看懂~ //
The number of features to draw from X to train each base estimator.
- If int, then draw max_features features.
- If float, then draw max_features * X.shape[1] features.
bootstrap:bool, optional (default=False)
//subsample时,采取有放回抽样还是无放回,默认不放回 //
If True, individual trees are fit on random subsets of the training data sampled with replacement. If False, sampling without replacement is performed.
n_jobs:int or None, optional (default=None)
//模型训练和预测时,工作的core数量,-1则全部用来工作//
The number of jobs to run in parallel for both fit and predict. None means 1 unless in a joblib.parallel_backend context. -1 means using all processors. See Glossary for more details.
random_state:int, RandomState instance or None, optional (default=None)
//设置随机数,方便结果可复现//
- If int, random_state is the seed used by the random number generator
- If RandomState instance, random_state is the random number generator
- If None, the random number generator is the RandomState instance used by np.random
verbose:int, optional (default=0)
//树的构建过程是否输出??//
Controls the verbosity of the tree building process.
warm_start:bool, optional (default=False)
//参考这里吧,我是没看懂 https://scikit-learn.org/stable/glossary.html#term-warm-start//
When set to True, reuse the solution of the previous call to fit and add more estimators to the ensemble, otherwise, just fit a whole new forest. See the Glossary.
Attributes
estimators_:list of DecisionTreeClassifier
The collection of fitted sub-estimators.
estimators_samples_:list of arrays
The subset of drawn samples for each base estimator.
max_samples_:integer
The actual number of samples
offset_:float
Offset used to define the decision function from the raw scores. We have the relation: decision_function = score_samples - offset_. offset_ is defined as follows. When the contamination parameter is set to “auto”, the offset is equal to -0.5 as the scores of inliers are close to 0 and the scores of outliers are close to -1. When a contamination parameter different than “auto” is provided, the offset is defined in such a way we obtain the expected number of outliers (samples with decision function < 0) in training.
Methods
////
decision_function(self, X)
//返回基学习器的平均得分=score_samples-offset;得分越低越不正常//
Average anomaly score of X of the base classifiers.
fit(self, X[, y, sample_weight])
//拟合模型//
Fit estimator.
fit_predict(self, X[, y])
//预测//
Perform fit on X and returns labels for X.
get_params(self[, deep])
//得到模型参数//
Get parameters for this estimator.
predict(self, X)
//预测//
Predict if a particular sample is an outlier or not.
score_samples(self, X)
//得分,计算逻辑得看下代码和paper//
Opposite of the anomaly score defined in the original paper.
set_params(self, **params)
//设置参数//
Set the parameters of this estimator.
Demo
Copy文档里简单的一维数据进行测试,查看各个方法输出的结果
import numpy as np
from sklearn.ensemble import IsolationForest
from sklearn.ensemble import _average_path_length
X = [[-1.1], [0.3], [0.5], [100]]
clf1 = IsolationForest(random_state=0)
clf1.fit(X)
clf2 = IsolationForest(random_state=0,contamination=0.1)
clf2.fit(X)
IsolationForest(behaviour='deprecated', bootstrap=False, contamination=0.1,
max_features=1.0, max_samples='auto', n_estimators=100,
n_jobs=None, random_state=0, verbose=0, warm_start=False)
模型的参数
clf1.get_params()
{'behaviour': 'deprecated',
'bootstrap': False,
'contamination': 'auto',
'max_features': 1.0,
'max_samples': 'auto',
'n_estimators': 100,
'n_jobs': None,
'random_state': 0,
'verbose': 0,
'warm_start': False}
预测得分
得分相关的方法有四个:
- _compute_score_samples: 返回结果记score1,和paper里score的计算逻辑一致
- _compute_chunked_score_samples:返回结果记score2,和paper里score的计算逻辑一致,且score1=score2
- score_samples:返回结果记score3,对_compute_score_samples结果取相反数,即-score3=score2=score1
- decision_function:score_samples返回结果-offset,其中offset默认为-0.5,如果contamination为float,比如0.1,那么offset为score3的10%分位数
clf1._compute_score_samples(np.asarray(X),False)
array([0.46946339, 0.32529681, 0.33144271, 0.68006339])
clf1._compute_chunked_score_samples(np.asarray(X))
array([0.46946339, 0.32529681, 0.33144271, 0.68006339])
clf1.score_samples(X)
array([-0.46946339, -0.32529681, -0.33144271, -0.68006339])
clf1.decision_function(X)
array([ 0.03053661, 0.17470319, 0.16855729, -0.18006339])
clf1.score_samples(X)-(-0.5)
array([ 0.03053661, 0.17470319, 0.16855729, -0.18006339])
预测标签
打标逻辑:decision_function小于0的就是异常样本
clf1.predict(X)
array([ 1, 1, 1, -1])
offset计算逻辑
offset和contamination参数有关,"auto"是为-0.5,否则通过float类型计算分位数
clf2.offset_
-0.6168833934367299
np.percentile(clf2.score_samples(X),100. *0.1)
-0.6168833934367299
Case 1
构造数据
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import IsolationForest
# 随机数相关,后期用来构造测试数据
rng = np.random.RandomState(42)
# Generate train data
# 生成100行2列array的0-1随机数,并且乘0.3,映射值域到[0,0.3]
X = 0.3 * rng.randn(100, 2)
# 对X分别加减2,得到两个(100,2)的矩阵
# 通过np.r_,对两个矩阵行合并,类似R里的rbind
X_train = np.r_[X + 2, X - 2]
# Generate some regular novel observations
# 下面生成测试数据,分布和生成逻辑保持一致
X1 = 0.3 * rng.randn(20, 2)
X_test = np.r_[X1 + 2, X1 - 2]
# Generate some abnormal novel observations
# 异常值由均匀分布U(-4,4)构造,20行2列的array
X_outliers = rng.uniform(low=-4, high=4, size=(20, 2))
模型拟合
# fit the model
clf = IsolationForest(max_samples=100, random_state=rng)
clf.fit(X_train)
y_pred_train = clf.predict(X_train)
y_pred_test = clf.predict(X_test)
y_pred_outliers = clf.predict(X_outliers)
查看结果
print("------ _compute_score_samples ------"+"\n");print(clf._compute_score_samples(np.asarray(X),False)[1:10])
print("------ _compute_chunked_score_samples ------"+"\n");print(clf._compute_chunked_score_samples(np.asarray(X))[1:10])
print("------ score_samples ------"+"\n");print(clf.score_samples(X)[1:10])
print("------ decision_function ------"+"\n");print(clf.decision_function(X)[1:10])
------ _compute_score_samples ------
[0.66061839 0.65897815 0.65788691 0.66281174 0.65897815 0.66289298
0.66678221 0.66171416 0.66163282]
------ _compute_chunked_score_samples ------
[0.66061839 0.65897815 0.65788691 0.66281174 0.65897815 0.66289298
0.66678221 0.66171416 0.66163282]
------ score_samples ------
[-0.66061839 -0.65897815 -0.65788691 -0.66281174 -0.65897815 -0.66289298
-0.66678221 -0.66171416 -0.66163282]
------ decision_function ------
[-0.16061839 -0.15897815 -0.15788691 -0.16281174 -0.15897815 -0.16289298
-0.16678221 -0.16171416 -0.16163282]
y_pred_train
array([ 1, -1, 1, -1, 1, 1, -1, -1, 1, -1, -1, 1, 1, 1, 1, -1, 1,
-1, -1, 1, 1, 1, -1, 1, -1, 1, 1, -1, 1, 1, 1, -1, -1, 1,
1, -1, 1, -1, 1, -1, 1, -1, 1, 1, 1, 1, 1, -1, 1, 1, 1,
1, 1, -1, 1, -1, -1, 1, 1, -1, 1, -1, -1, 1, 1, -1, 1, -1,
1, -1, 1, -1, 1, -1, 1, 1, 1, 1, -1, 1, 1, -1, -1, -1, 1,
1, 1, 1, 1, -1, 1, 1, 1, 1, -1, 1, 1, 1, 1, 1, 1, -1,
1, -1, 1, 1, -1, -1, 1, -1, -1, -1, 1, 1, 1, -1, 1, -1, -1,
-1, 1, 1, -1, 1, -1, 1, 1, -1, 1, 1, 1, -1, -1, 1, 1, -1,
-1, -1, 1, -1, 1, -1, 1, 1, 1, 1, 1, -1, 1, 1, -1, 1, 1,
-1, 1, -1, -1, 1, 1, -1, 1, -1, -1, -1, 1, -1, 1, -1, 1, -1,
1, -1, 1, -1, 1, 1, 1, 1, -1, -1, 1, -1, -1, -1, 1, -1, 1,
1, -1, -1, 1, 1, 1, 1, -1, 1, 1, 1, 1, 1])
可视化
# plot the line, the samples, and the nearest vectors to the plane
#
xx, yy = np.meshgrid(np.linspace(-5, 5, 50), np.linspace(-5, 5, 50))
# xx.ravel()转成1维
# np.c_列拼接cbind
Z = clf.decision_function(np.c_[xx.ravel(), yy.ravel()])
# 重塑
Z = Z.reshape(xx.shape)
plt.title("IsolationForest")
plt.contourf(xx, yy, Z, cmap=plt.cm.Blues_r)
# 白色的点为train
b1 = plt.scatter(X_train[:, 0], X_train[:, 1], c='white',
s=20, edgecolor='k')
# 绿色的点为test
b2 = plt.scatter(X_test[:, 0], X_test[:, 1], c='yellow',
s=20, edgecolor='k')
# 红色点为outliers
c = plt.scatter(X_outliers[:, 0], X_outliers[:, 1], c='red',
s=20, edgecolor='k')
plt.axis('tight')
plt.xlim((-5, 5))
plt.ylim((-5, 5))
plt.legend([b1, b2, c],
["training observations",
"new regular observations", "new abnormal observations"],
loc="upper left")
plt.show()
从上面的图来看,红色的异常点,确实大部分分布在簇之外,但是如果从预测的结果看,簇中很多点,也被标记为异常值.总之,感觉iforest的说服力不是很够,最终还是需要人工check,不知道在业界是怎么玩滴。
https://scikit-learn.org/stable/auto_examples/plot_anomaly_comparison.html#sphx-glr-auto-examples-plot-anomaly-comparison-py介绍了多种算法的比较可以再关注下,或者Paper里用多个数据,通过AUC这个指标进行测试,也可以复现下试试看
Case2-Credit Card Fraud Detection
Intro
使用kaggle的信用卡盗刷数据,检验iForest效果。数据为信用卡的交易记录,共计284,807条,其中492条盗刷行为,占比0.172%。提供的数据是脱敏之后的,由pca提供的数据,全部都是数值型,也没有缺失值,很适合做无监督的测试。
不同算法效果比较,通过5折交叉验证,采用AUC,其余指标和阈值有关系,不适合直接比较。为了保证结果可复现,需要控制两个随机性:
- 5折交叉验证时,数据集划分的随机性
- 模型训练时的随机性
EDA
简单的EDA,对数据做简单探索。
import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings('ignore')
rawdata = pd.read_csv("../Data/creditcard.csv")
rawdata.head()
| Time | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | ... | V21 | V22 | V23 | V24 | V25 | V26 | V27 | V28 | Amount | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | -1.359807 | -0.072781 | 2.536347 | 1.378155 | -0.338321 | 0.462388 | 0.239599 | 0.098698 | 0.363787 | ... | -0.018307 | 0.277838 | -0.110474 | 0.066928 | 0.128539 | -0.189115 | 0.133558 | -0.021053 | 149.62 | 0 |
| 1 | 0.0 | 1.191857 | 0.266151 | 0.166480 | 0.448154 | 0.060018 | -0.082361 | -0.078803 | 0.085102 | -0.255425 | ... | -0.225775 | -0.638672 | 0.101288 | -0.339846 | 0.167170 | 0.125895 | -0.008983 | 0.014724 | 2.69 | 0 |
| 2 | 1.0 | -1.358354 | -1.340163 | 1.773209 | 0.379780 | -0.503198 | 1.800499 | 0.791461 | 0.247676 | -1.514654 | ... | 0.247998 | 0.771679 | 0.909412 | -0.689281 | -0.327642 | -0.139097 | -0.055353 | -0.059752 | 378.66 | 0 |
| 3 | 1.0 | -0.966272 | -0.185226 | 1.792993 | -0.863291 | -0.010309 | 1.247203 | 0.237609 | 0.377436 | -1.387024 | ... | -0.108300 | 0.005274 | -0.190321 | -1.175575 | 0.647376 | -0.221929 | 0.062723 | 0.061458 | 123.50 | 0 |
| 4 | 2.0 | -1.158233 | 0.877737 | 1.548718 | 0.403034 | -0.407193 | 0.095921 | 0.592941 | -0.270533 | 0.817739 | ... | -0.009431 | 0.798278 | -0.137458 | 0.141267 | -0.206010 | 0.502292 | 0.219422 | 0.215153 | 69.99 | 0 |
5 rows × 31 columns
rawdata.shape
(284807, 31)
rawdata.Class.value_counts()
0 284315
1 492
Name: Class, dtype: int64
rawdata.isna().sum()
Time 0
V1 0
V2 0
V3 0
V4 0
V5 0
V6 0
V7 0
V8 0
V9 0
V10 0
V11 0
V12 0
V13 0
V14 0
V15 0
V16 0
V17 0
V18 0
V19 0
V20 0
V21 0
V22 0
V23 0
V24 0
V25 0
V26 0
V27 0
V28 0
Amount 0
Class 0
dtype: int64
- 数据维度:284807行,31列
- 正负样本分布:0-284315;1-492
- 所有列均无缺失值
不做其他数据探索,直接上算法
数据准备
为了保证数据可比和可重复,交叉验证的数据划分和模型训练需要指定随机数种子。
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV,StratifiedKFold
rawdata['Hour'] =rawdata["Time"].apply(lambda x : divmod(x, 3600)[0])
# 特征数据单独存放
X=rawdata.drop(['Time','Class'],axis=1)
# label数据单独存放
y=rawdata.Class
X.shape
(284807, 30)
训练、测试数据划分
调用train_test_split函数划分训练集和测试集,设置参数stratify=y,保证划分之后正负样本比和总体样本保持一致。
train_test_split用法 https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.train_test_split.html#sklearn.model_selection.train_test_split
X_traintotal,X_test,y_traintotal,y_test=train_test_split(X,y,stratify=y,test_size=0.2,random_state =12345)
y_traintotal.value_counts()
0 227451
1 394
Name: Class, dtype: int64
y_test.value_counts()
0 56864
1 98
Name: Class, dtype: int64
5折划分
StratifiedKFold同样可以保证各个折的数据中,正负样本分布和总体一致
StratifiedKFold https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.StratifiedKFold.html#sklearn.model_selection.StratifiedKFold
splits为生成器,只能访问一次。
//假如我们要生成从 1 到 10 这 10 个数字,采用列表的方式定义,会占用 10 个地址空间。采用生成器,只会占用一个地址空间。因为生成器并没有把所有的值存在内存中,而是在运行时生成值。所以生成器只能访问一次。//
NFOLDS = 5
folds = StratifiedKFold(n_splits=NFOLDS,random_state=12345)
splits = folds.split(X_traintotal, y_traintotal)
enumerate函数用法:https://www.runoob.com/python/python-func-enumerate.html
就是给可遍历的数据对象组合成一个索引序列
新建一个list,把K折数据拼接成字典放进去
kFoldsList = []
for fold_n, (train_index, valid_index) in enumerate(splits):
kFoldsList.append({
"fold": fold_n,
"train_index": train_index,
"valid_index": valid_index
})
kFoldsList
[{'fold': 0,
'train_index': array([ 40589, 41263, 41754, ..., 227842, 227843, 227844]),
'valid_index': array([ 0, 1, 2, ..., 45571, 45572, 45573])},
{'fold': 1,
'train_index': array([ 0, 1, 2, ..., 227842, 227843, 227844]),
'valid_index': array([40589, 41263, 41754, ..., 91141, 91142, 91143])},
{'fold': 2,
'train_index': array([ 0, 1, 2, ..., 227842, 227843, 227844]),
'valid_index': array([ 87243, 87254, 87884, ..., 136716, 136717, 136718])},
{'fold': 3,
'train_index': array([ 0, 1, 2, ..., 227842, 227843, 227844]),
'valid_index': array([133467, 133512, 134189, ..., 182282, 182283, 182284])},
{'fold': 4,
'train_index': array([ 0, 1, 2, ..., 182282, 182283, 182284]),
'valid_index': array([177302, 177309, 178759, ..., 227842, 227843, 227844])}]
查看下正负样本比
for i in range(len(kFoldsList)):
print("--- 第"+str(i)+"折数据 ---")
validY = y_traintotal[kFoldsList[i]["valid_index"]]
print(" --- 正负样本比: ",str(validY.sum()/validY.count()))
--- 第0折数据 ---
--- 正负样本比: 0.003181219833260202
--- 第1折数据 ---
--- 正负样本比: 0.0014567242943132781
--- 第2折数据 ---
--- 正负样本比: 0.0009863824423925254
--- 第3折数据 ---
--- 正负样本比: 0.0023323455164087365
--- 第4折数据 ---
--- 正负样本比: 0.0011241808560225933
下面查看k折的效果,主要关注以下几个方便:
- valid折数据拼起来是否是原始数据的全集
- 每一折数据的train和valid是否都没有交集且并集是原始数据的全集
for i in range(len(kFoldsList)):
print("------ 第%d折验证集索引 ------"%i)
print(" --- top3: %s"% str(kFoldsList[i]["valid_index"][0:3]))
print(" --- tail3: %s"% str(kFoldsList[i]["valid_index"][-3:]))
------ 第0折验证集索引 ------
--- top3: [0 1 2]
--- tail3: [45571 45572 45573]
------ 第1折验证集索引 ------
--- top3: [40589 41263 41754]
--- tail3: [91141 91142 91143]
------ 第2折验证集索引 ------
--- top3: [87243 87254 87884]
--- tail3: [136716 136717 136718]
------ 第3折验证集索引 ------
--- top3: [133467 133512 134189]
--- tail3: [182282 182283 182284]
------ 第4折验证集索引 ------
--- top3: [177302 177309 178759]
--- tail3: [227842 227843 227844]
validTestList = []
for i in kFoldsList:
validTestList.extend(i["valid_index"])
set(range(len(X_traintotal)))-set(validTestList)
set()
数据没有问题
LR Baseline
训练LR模型,和iForest比较。采用5折交叉验证,比较指标选取AUC。
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import roc_auc_score, roc_curve, precision_score, auc, precision_recall_curve, accuracy_score, recall_score, f1_score, confusion_matrix, classification_report
columns = X_traintotal.columns
y_preds = np.zeros(X_test.shape[0])
# 类似rf的带外误差估计,把valid的那一折数据放在这个变量中
y_oof = np.zeros(X_traintotal.shape[0])
score = 0
for fold_n, itemDict in enumerate(kFoldsList):
train_index =itemDict["train_index"]
valid_index=itemDict["valid_index"]
X_train, X_valid = X_traintotal[columns].iloc[train_index], X_traintotal[columns].iloc[valid_index]
y_train, y_valid = y_traintotal.iloc[train_index], y_traintotal.iloc[valid_index]
clf = LogisticRegression(random_state=123,n_jobs=2)
clf.fit(X_train,y_train)
y_pred_valid = clf.predict_proba(X_valid)[:,1]
# 把交叉验证的结果保存在y_oof中
y_oof[valid_index] = y_pred_valid
print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf.predict_proba(X_test)[:,1] / NFOLDS
del X_train, X_valid, y_train, y_valid
print(f"\nMean AUC = {score}")
print(f"Out of folds AUC = {roc_auc_score(y_traintotal, y_oof)}")
print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
Fold 1 | AUC: 0.9654628782588159
Fold 2 | AUC: 0.9791964293167785
Fold 3 | AUC: 0.9245720995851086
Fold 4 | AUC: 0.9615806506368071
Fold 5 | AUC: 0.9636139254419527
Mean AUC = 0.9588851966478926
Out of folds AUC = 0.958075881217859
test datasets AUC = 0.9449782797193159
LR的auc就很高,数据可能本身的可分性就很好
iForest
测试函数封装
from sklearn.ensemble import IsolationForest
def iForestTest(model, kFoldsList=kFoldsList,X_traintotal=X_traintotal,y_traintotal=y_traintotal,X_test=X_test,y_test=y_test):
columns = X_traintotal.columns
y_preds = np.zeros(X_test.shape[0])
# 类似rf的带外误差估计,把valid的那一折数据放在这个变量中
y_oof = np.zeros(X_traintotal.shape[0])
score = 0
for fold_n, itemDict in enumerate(kFoldsList):
train_index =itemDict["train_index"]
valid_index=itemDict["valid_index"]
X_train, X_valid = X_traintotal[columns].iloc[train_index], X_traintotal[columns].iloc[valid_index]
y_train, y_valid = y_traintotal.iloc[train_index], y_traintotal.iloc[valid_index]
# clf = IsolationForest(n_estimators=100,n_jobs=1,random_state=123,verbose=0)
clf = model
clf.fit(X_train)
y_pred_valid = clf._compute_score_samples(X_valid, False)
y_oof[valid_index] = y_pred_valid
# print(f"Fold {fold_n + 1} | AUC: {roc_auc_score(y_valid, y_pred_valid)}")
score += roc_auc_score(y_valid, y_pred_valid) / NFOLDS
y_preds += clf._compute_score_samples(X_test, False) / NFOLDS
# print(f"\nMean AUC = {score}")
# 这个值和score的差别很小,相当于一个是计算全集的auc,另一个是把全集分成5个部分,算这5个部分的平均auc
# print(f"Out of folds AUC = {roc_auc_score(y_traintotal, y_oof)}")
# print(f"test datasets AUC = {roc_auc_score(y_test, y_preds)}")
return score,roc_auc_score(y_test, y_preds)
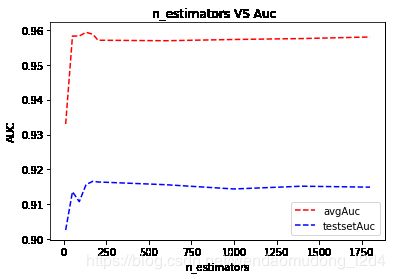
n_estimators VS auc
list(range(10,200,40))+list(range(200,2100,400))
[10, 50, 90, 130, 170, 200, 600, 1000, 1400, 1800]
nEstimatorsNum = []
kFlodsAvgAuc = []
testSetAuc = []
for i in list(range(10,200,40))+list(range(200,2100,400)):
print("------- 参数=",str(i))
iForestModel = IsolationForest(n_estimators=i,max_samples=256,n_jobs=-1,random_state=123,verbose=0)
result = iForestTest(model=iForestModel, kFoldsList=kFoldsList,X_traintotal=X_traintotal,X_test=X_test,y_test=y_test)
nEstimatorsNum.append(i)
kFlodsAvgAuc.append(result[0])
testSetAuc.append(result[1])
------- 参数= 10
------- 参数= 50
------- 参数= 90
------- 参数= 130
------- 参数= 170
------- 参数= 200
------- 参数= 600
------- 参数= 1000
------- 参数= 1400
------- 参数= 1800
df1 = pd.DataFrame({"nEstimatorsNum":nEstimatorsNum,"kFlodsAvgAuc":kFlodsAvgAuc,"testSetAuc":testSetAuc})
df1
| nEstimatorsNum | kFlodsAvgAuc | testSetAuc | |
|---|---|---|---|
| 0 | 10 | 0.933037 | 0.902601 |
| 1 | 50 | 0.958268 | 0.913583 |
| 2 | 90 | 0.958288 | 0.910744 |
| 3 | 130 | 0.959342 | 0.915648 |
| 4 | 170 | 0.958792 | 0.916589 |
| 5 | 200 | 0.957079 | 0.916391 |
| 6 | 600 | 0.956933 | 0.915600 |
| 7 | 1000 | 0.957292 | 0.914381 |
| 8 | 1400 | 0.957555 | 0.915176 |
| 9 | 1800 | 0.958007 | 0.914904 |
import matplotlib.pyplot as plt
plt.plot(df1.nEstimatorsNum,df1.kFlodsAvgAuc,'r--',label='avgAuc')
plt.plot(df1.nEstimatorsNum,df1.testSetAuc,'b--',label='testsetAuc')
plt.title("n_estimators VS Auc ")
plt.xlabel('n_estimators')
plt.ylabel('AUC')
plt.legend()
就该数据而言:
- n_estimators=130时,auc指标达到最优
- 后续增加n_estimators,auc提升不明显
max_samples VS auc
maxSamplesNum = []
kFlodsAvgAuc1 = []
testSetAuc1 = []
for i in list(range(10,300,20))+list(range(200,2100,400)):
print("------- 参数maxSamples=",str(i))
iForestModel = IsolationForest(n_estimators=130,max_samples=i,n_jobs=-1,random_state=123,verbose=0)
result = iForestTest(model=iForestModel, kFoldsList=kFoldsList,X_traintotal=X_traintotal,X_test=X_test,y_test=y_test)
maxSamplesNum.append(i)
kFlodsAvgAuc1.append(result[0])
testSetAuc1.append(result[1])
------- 参数maxSamples= 10
------- 参数maxSamples= 30
------- 参数maxSamples= 50
------- 参数maxSamples= 70
------- 参数maxSamples= 90
------- 参数maxSamples= 110
------- 参数maxSamples= 130
------- 参数maxSamples= 150
------- 参数maxSamples= 170
------- 参数maxSamples= 190
------- 参数maxSamples= 210
------- 参数maxSamples= 230
------- 参数maxSamples= 250
------- 参数maxSamples= 270
------- 参数maxSamples= 290
------- 参数maxSamples= 200
------- 参数maxSamples= 600
------- 参数maxSamples= 1000
------- 参数maxSamples= 1400
------- 参数maxSamples= 1800
df2 = pd.DataFrame({"maxSamplesNum":maxSamplesNum,"kFlodsAvgAuc":kFlodsAvgAuc1,"testSetAuc":testSetAuc1})
df2
| maxSamplesNum | kFlodsAvgAuc | testSetAuc | |
|---|---|---|---|
| 0 | 10 | 0.949487 | 0.903520 |
| 1 | 30 | 0.951984 | 0.906453 |
| 2 | 50 | 0.953766 | 0.908032 |
| 3 | 70 | 0.958786 | 0.909094 |
| 4 | 90 | 0.959577 | 0.914187 |
| 5 | 110 | 0.957167 | 0.912080 |
| 6 | 130 | 0.959681 | 0.911190 |
| 7 | 150 | 0.958994 | 0.913248 |
| 8 | 170 | 0.959381 | 0.915583 |
| 9 | 190 | 0.959911 | 0.914841 |
| 10 | 210 | 0.958282 | 0.914825 |
| 11 | 230 | 0.958917 | 0.912698 |
| 12 | 250 | 0.959981 | 0.916094 |
| 13 | 270 | 0.960119 | 0.915770 |
| 14 | 290 | 0.960576 | 0.914974 |
| 15 | 200 | 0.958839 | 0.914398 |
| 16 | 600 | 0.959962 | 0.919391 |
| 17 | 1000 | 0.960982 | 0.917315 |
| 18 | 1400 | 0.961815 | 0.917679 |
| 19 | 1800 | 0.960359 | 0.916489 |
plt.plot(df2.maxSamplesNum,df2.kFlodsAvgAuc,'r--',label='avgAuc')
plt.plot(df2.maxSamplesNum,df2.testSetAuc,'b--',label='testsetAuc')
plt.title("maxSamples VS Auc ")
plt.xlabel('maxSamples')
plt.ylabel('AUC')
plt.legend()
contamination对算法效果的影响
contamination和阈值紧密相关,测试不同contamination对precision、recall的影响,方便起见,只观测测试集的效果
y_traintotal.value_counts()
0 227451
1 394
Name: Class, dtype: int64
394/(394+227451)
0.001729245759178389
from sklearn import metrics
for i in [0.0017,0.005,0.01,0.05,0.1]:
print("---------- contamination: "+str(i))
iForestModel = IsolationForest(n_estimators=100,max_samples=256,n_jobs=-1,random_state=123,verbose=0,contamination=i)
iForestModel.fit(X_traintotal)
test_Pre = iForestModel.predict(X_test)
test_Pre1 = [1 if i==-1 else 0 for i in list(test_Pre)]
#--- report
print('\n',metrics.classification_report(y_test, test_Pre1))
---------- contamination: 0.0017
precision recall f1-score support
0 1.00 1.00 1.00 56864
1 0.36 0.31 0.33 98
accuracy 1.00 56962
macro avg 0.68 0.65 0.66 56962
weighted avg 1.00 1.00 1.00 56962
---------- contamination: 0.005
precision recall f1-score support
0 1.00 1.00 1.00 56864
1 0.16 0.45 0.24 98
accuracy 0.99 56962
macro avg 0.58 0.72 0.62 56962
weighted avg 1.00 0.99 1.00 56962
---------- contamination: 0.01
precision recall f1-score support
0 1.00 0.99 1.00 56864
1 0.10 0.56 0.17 98
accuracy 0.99 56962
macro avg 0.55 0.78 0.58 56962
weighted avg 1.00 0.99 0.99 56962
---------- contamination: 0.05
precision recall f1-score support
0 1.00 0.95 0.98 56864
1 0.03 0.76 0.05 98
accuracy 0.95 56962
macro avg 0.51 0.85 0.51 56962
weighted avg 1.00 0.95 0.97 56962
---------- contamination: 0.1
precision recall f1-score support
0 1.00 0.90 0.95 56864
1 0.01 0.82 0.03 98
accuracy 0.90 56962
macro avg 0.51 0.86 0.49 56962
weighted avg 1.00 0.90 0.95 56962
Summary
就这个数据集而言:
- 训练集上LR和iForest表现差不多,测试集上LR较优
- 调参对iForest算法提升有限
- 实际应用时,最好不要根据阈值划分,而是去score top数据做验证,归纳规则再应用到实际业务中
Ref
[1] Sklearn文档 https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.IsolationForest.html
[2] Case1文档 https://scikit-learn.org/stable/auto_examples/ensemble/plot_isolation_forest.html#sphx-glr-auto-examples-ensemble-plot-isolation-forest-py
[3] Case2背景 https://www.kaggle.com/mlg-ulb/creditcardfraud
[4] https://zhuanlan.zhihu.com/p/93779599
2020-01-14 于南京市江宁区九龙湖