【keras框架下Resnet101_Unet深度学习模型对医学核磁图像语义分割】
盆骨分割模型
前言
U-Net和FCN非常的相似,U-Net比FCN稍晚提出来,但都发表在2015年,和FCN相比,U-Net的第一个特点是完全对称,也就是左边和右边是很类似的,而FCN的decoder相对简单,只用了一个deconvolution的操作,之后并没有跟上卷积结构。第二个区别就是skip connection,FCN用的是加操作(summation),U-Net用的是叠操作(concatenation)。这些都是细节,重点是它们的结构用了一个比较经典的思路,也就是编码和解码(encoder-decoder),早在2006年就被Hinton大神提出来发表在了nature上.
这个网红结构,我们先提取出它的拓扑结构,这样会比较容易分析它的实质,排除很多细节的干扰。输入是一幅图,输出是目标的分割结果。继续简化就是,一幅图,编码,或者说降采样,然后解码,也就是升采样,然后输出一个分割结果。根据结果和真实分割的差异,反向传播来训练这个分割网络。我们可以说,U-Net里面最精彩的部分就是这三部分:
- 下采样
- 上采样
- skip connection
拿U-Net来说,原论文给出的结构是原图经过四次降采样,四次上采样,得到分割结果,实际呢,为什么四次?就是作者喜欢呗,或者说当时作者使用的数据集,四次降采样的效果好;Unet中对于需要多深的问题。其实这个是非常灵活的,涉及到的一个点就是特征提取器,各种在encoder上的微创新络绎不绝,最直接的就是用ImageNet里面的明星结构来套嘛,前几年的BottleNeck,VGG16,Residual,还有去年的DenseNet,就比谁出文章快。这一类的论文就相当于从1到10的递进,而U-Net这个低层结构的提出却是从0到1。说特征提取器是dense block,名字也就是DenseUNet,或者是residual block效果好,然后名字也就是ResUNet。其他的大家可以看这篇文章:https://zhuanlan.zhihu.com/p/44958351
keras中如何定义残差网络:https://blog.csdn.net/Tourior/article/details/83824436
1说明
本次采用的是vnet2d二分类模型和Resnet101_Unet模型分别进行训练测试,为防止样本分布不均衡现象,损失函数均采用二分类dice损失,不再使用之前的最原始的二分类交叉熵损失。
二分类dice损失如下:
Resnet101_Unet模型实现如下,值得注意的是原始Unet网络相对交浅,特征提取能力有限,故这里我们使用Resnet101进行网络模型加深,提高特征提取效果。
# *******************resnet101 unet*********************
def conv3x3(x, out_filters, strides=(1, 1)):
x = Conv2D(out_filters, 3, padding='same', strides=strides, use_bias=False, kernel_initializer='he_normal')(x)
return x
def Conv2d_BN(x, nb_filter, kernel_size, strides=(1, 1), padding='same', use_activation=True):
x = Conv2D(nb_filter, kernel_size, padding=padding, strides=strides, kernel_initializer='he_normal')(x)
x = BatchNormalization(axis=3)(x)
if use_activation:
x = Activation('relu')(x)
return x
else:
return x
def basic_Block(input, out_filters, strides=(1, 1), with_conv_shortcut=False):
x = conv3x3(input, out_filters, strides)
x = BatchNormalization(axis=3)(x)
x = Activation('relu')(x)
x = conv3x3(x, out_filters)
x = BatchNormalization(axis=3)(x)
if with_conv_shortcut:
residual = Conv2D(out_filters, 1, strides=strides, use_bias=False, kernel_initializer='he_normal')(input)
residual = BatchNormalization(axis=3)(residual)
x = add([x, residual])
else:
x = add([x, input])
x = Activation('relu')(x)
return x
def bottleneck_Block(input, out_filters, strides=(1, 1), with_conv_shortcut=False):
expansion = 4
de_filters = int(out_filters / expansion)
x = Conv2D(de_filters, 1, use_bias=False, kernel_initializer='he_normal')(input)
x = BatchNormalization(axis=3)(x)
x = Activation('relu')(x)
x = Conv2D(de_filters, 3, strides=strides, padding='same', use_bias=False, kernel_initializer='he_normal')(x)
x = BatchNormalization(axis=3)(x)
x = Activation('relu')(x)
x = Conv2D(out_filters, 1, use_bias=False, kernel_initializer='he_normal')(x)
x = BatchNormalization(axis=3)(x)
if with_conv_shortcut:
residual = Conv2D(out_filters, 1, strides=strides, use_bias=False, kernel_initializer='he_normal')(input)
residual = BatchNormalization(axis=3)(residual)
x = add([x, residual])
else:
x = add([x, input])
x = Activation('relu')(x)
return x
def unet_resnet_101(height=320, width=320, channel=1, classes=3):
input = Input(shape=(height, width, channel))
conv1_1 = Conv2D(64, 7, strides=(2, 2), padding='same', use_bias=False, kernel_initializer='he_normal')(input)
conv1_1 = BatchNormalization(axis=3)(conv1_1)
conv1_1 = Activation('relu')(conv1_1)
conv1_2 = MaxPooling2D(pool_size=(3, 3), strides=(2, 2), padding='same')(conv1_1)
# conv2_x 1/4
conv2_1 = bottleneck_Block(conv1_2, 256, strides=(1, 1), with_conv_shortcut=True)
conv2_2 = bottleneck_Block(conv2_1, 256)
conv2_3 = bottleneck_Block(conv2_2, 256)
# conv3_x 1/8
conv3_1 = bottleneck_Block(conv2_3, 512, strides=(2, 2), with_conv_shortcut=True)
conv3_2 = bottleneck_Block(conv3_1, 512)
conv3_3 = bottleneck_Block(conv3_2, 512)
conv3_4 = bottleneck_Block(conv3_3, 512)
# conv4_x 1/16
conv4_1 = bottleneck_Block(conv3_4, 1024, strides=(2, 2), with_conv_shortcut=True)
conv4_2 = bottleneck_Block(conv4_1, 1024)
conv4_3 = bottleneck_Block(conv4_2, 1024)
conv4_4 = bottleneck_Block(conv4_3, 1024)
conv4_5 = bottleneck_Block(conv4_4, 1024)
conv4_6 = bottleneck_Block(conv4_5, 1024)
conv4_7 = bottleneck_Block(conv4_6, 1024)
conv4_8 = bottleneck_Block(conv4_7, 1024)
conv4_9 = bottleneck_Block(conv4_8, 1024)
conv4_10 = bottleneck_Block(conv4_9, 1024)
conv4_11 = bottleneck_Block(conv4_10, 1024)
conv4_12 = bottleneck_Block(conv4_11, 1024)
conv4_13 = bottleneck_Block(conv4_12, 1024)
conv4_14 = bottleneck_Block(conv4_13, 1024)
conv4_15 = bottleneck_Block(conv4_14, 1024)
conv4_16 = bottleneck_Block(conv4_15, 1024)
conv4_17 = bottleneck_Block(conv4_16, 1024)
conv4_18 = bottleneck_Block(conv4_17, 1024)
conv4_19 = bottleneck_Block(conv4_18, 1024)
conv4_20 = bottleneck_Block(conv4_19, 1024)
conv4_21 = bottleneck_Block(conv4_20, 1024)
conv4_22 = bottleneck_Block(conv4_21, 1024)
conv4_23 = bottleneck_Block(conv4_22, 1024)
# conv5_x 1/32
conv5_1 = bottleneck_Block(conv4_23, 2048, strides=(2, 2), with_conv_shortcut=True)
conv5_2 = bottleneck_Block(conv5_1, 2048)
conv5_3 = bottleneck_Block(conv5_2, 2048)
up6 = Conv2d_BN(UpSampling2D(size=(2, 2))(conv5_3), 1024, 2)
merge6 = concatenate([conv4_23, up6], axis=3)
conv6 = Conv2d_BN(merge6, 1024, 3)
conv6 = Conv2d_BN(conv6, 1024, 3)
up7 = Conv2d_BN(UpSampling2D(size=(2, 2))(conv6), 512, 2)
merge7 = concatenate([conv3_4, up7], axis=3)
conv7 = Conv2d_BN(merge7, 512, 3)
conv7 = Conv2d_BN(conv7, 512, 3)
up8 = Conv2d_BN(UpSampling2D(size=(2, 2))(conv7), 256, 2)
merge8 = concatenate([conv2_3, up8], axis=3)
conv8 = Conv2d_BN(merge8, 256, 3)
conv8 = Conv2d_BN(conv8, 256, 3)
up9 = Conv2d_BN(UpSampling2D(size=(2, 2))(conv8), 64, 2)
merge9 = concatenate([conv1_1, up9], axis=3)
conv9 = Conv2d_BN(merge9, 64, 3)
conv9 = Conv2d_BN(conv9, 64, 3)
up10 = Conv2d_BN(UpSampling2D(size=(2, 2))(conv9), 64, 2)
conv10 = Conv2d_BN(up10, 64, 3)
conv10 = Conv2d_BN(conv10, 64, 3)
conv11 = Conv2d_BN(conv10, classes, 1, use_activation=None)
activation = Activation('softmax', name='Classification')(conv11)
# conv_out=Conv2D(classes, 1, activation = 'softmax', padding = 'same', kernel_initializer = 'he_normal')(conv11)
model = Model(inputs=input, outputs=activation)
# print(model.output_shape) compounded_loss
# model_dice=dice_p_bce
# model_dice=compounded_loss(smooth=0.0005,gamma=2., alpha=0.25)
# model_dice=tversky_coef_loss_fun(alpha=0.3,beta=0.7)
# model_dice=dice_coef_loss_fun(smooth=1e-5)
# model.compile(optimizer = Nadam(lr = 2e-4), loss = model_dice, metrics = ['accuracy'])
#不使用metric
# model_dice=focal_loss(alpha=.25, gamma=2)
# model.compile(optimizer = Adam(lr = 2e-5),loss=dice_coef,metrics=['accuracy'])
model.compile(optimizer = Nadam(lr = 2e-5), loss = focal_lossm,metrics=['accuracy'])
# model.compile(optimizer = Nadam(lr = 2e-4), loss = "categorical_crossentropy",metrics=['accuracy'])
return model
核磁样本勾画33个共使用710个样本。盆骨基本每个层面都有,样本数量相对并不是很多,这里暂不进行数据集拓展,不使用验证集控制训练。
Resnet101_Unet模型训练参数如下表
|
|
batch_size |
epochs |
loss |
accurancy |
参数量(千万) |
| Vnet |
8 |
100 |
0.013 |
0.999 |
5.15 |
2训练过程
2.1 Resnet101_Unet模型
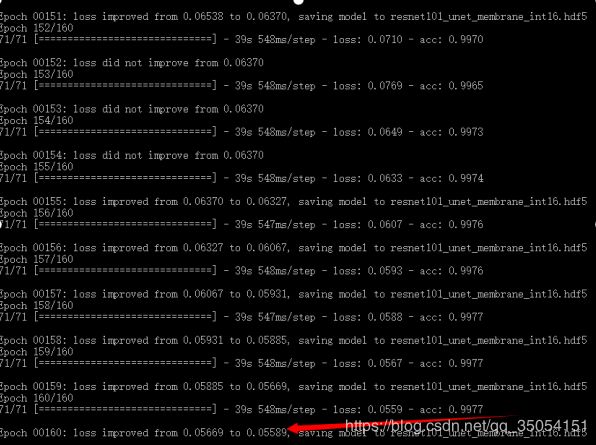
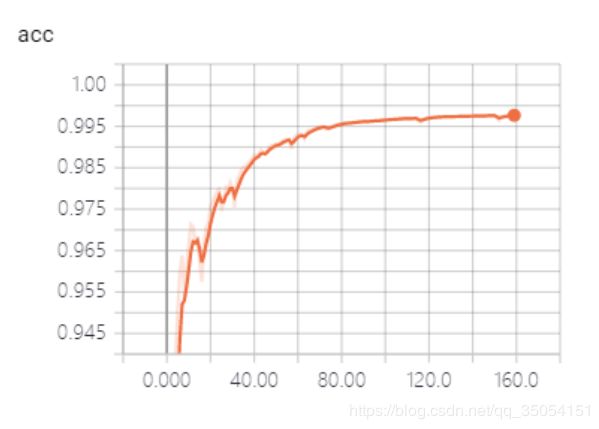
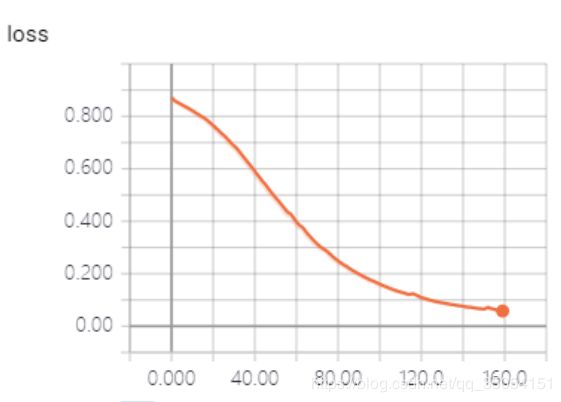
训练过程发现Resnet101_Unet模型可能因为待训练参数数量很大,如下图1显示Resnet101_Unet模型参数量将近9千万,比vnet2d网络5千万参数多了将近一倍,导致loss下降更新非常缓慢,在我个人本地主机训练将近10小时,loss呈现下降趋势但距离下降到目标期望范围花费时间较大。Resnet101_Unet模型同样采用dice损失函数训练100个epoch,batch_size为8.
图1 Unet模型训练截止图 图2 Unet模型参数量
图3 unet训练准确率曲线 图4 unet训练loss下降曲线
2.分割效果
测试选用12个未压脂核磁数据集,共计263个切片作为测试样本。测试集整体预测相对良好
3 下一步计划
1.损失函数改进:
本次训练采用DIce损失来应对样本不均衡现象,后续可采用generalized dice loss,tversky coefficient loss等新的损失或者Dice+交叉熵复合损失函数作为指导;另外针对样本集可采用改变对比度等方法适当增加拓展数据量,对测试分割较差的样本投入训练集。
2. 特征提取网络的改进:
目前采用的是Resnet101做骨干提取网络,后续打算测试Densenet网络。
训练集不开源,代码后续开源