【K-means算法】{1} —— 使用Python实现K-means算法并处理Iris数据集
此处基于K-means算法处理Iris数据集
Kmeans.py模块:
import numpy as np
class KMeansClassifier():
"""初始化KMeansClassifier类"""
def __init__(self, k=3, initCent='random', max_iter=500):
# 类的成员数据(变量前用下划线)
self._k = k # 中心点
self._initCent = initCent # 生成初始中心点
self._max_iter = max_iter # 最大迭代次数
self._clusterAssment = None # 点分配结果
self._labels = None
self._sse = None # 误差平方和
def _calEDist(self, arrA, arrB):
"""计算欧氏距离,参数为一维数组"""
return np.math.sqrt(sum(np.power(arrA-arrB, 2)))
def _calMDist(self, arrA, arrB):
"""计算曼哈顿距离,参数为一维数组"""
return sum(np.abs(arrA-arrB))
def _randCent(self, data_X, k):
"""随机选取k个质心,返回一个k*n的质心矩阵"""
n = data_X.shape[1] # 特征的维度
centroids = np.empty((k,n)) # 使用numpy生成一个k*n的矩阵,用于存储质心
for j in range(n):
minJ = min(data_X[:, j])
rangeJ = float(max(data_X[:, j] - minJ))
centroids[:, j] = (minJ + rangeJ * np.random.rand(k, 1)).flatten() # 使用flatten函数展平嵌套列表(nested list)
return centroids
def fit(self, data_X):
"""训练模型,参数为m*n维的矩阵"""
if not isinstance(data_X, np.ndarray):
data_X = np.asarray(data_X)
m = data_X.shape[0] # 样本的个数
self._clusterAssment = np.zeros((m,2)) # 一个m*2维矩阵,矩阵第一列存储样本点所属的族的索引值,第二列存储该点与所属族的质心的平方误差
if self._initCent == 'random':
self._centroids = self._randCent(data_X, self._k)
clusterChanged = True
for _ in range(self._max_iter):
clusterChanged = False
for i in range(m): # 将每个样本点分配到离它最近的质心所属的族
minDist = np.inf # 首先将minDist置为一个无穷大的数
minIndex = -1 # 将最近质心的下标置为-1
for j in range(self._k): # k次迭代用于寻找最近的质心
arrA = self._centroids[j,:]
arrB = data_X[i,:]
distJI = self._calEDist(arrA, arrB) # 计算误差值
if distJI < minDist:
minDist = distJI
minIndex = j
if self._clusterAssment[i, 0] != minIndex or self._clusterAssment[i, 1] > minDist**2:
clusterChanged = True
self._clusterAssment[i,:] = minIndex, minDist**2
if not clusterChanged: # 若所有样本点所属的族都不改变,则已收敛,结束迭代
break
# 更新质心,将每个族中的点的均值作为质心
for i in range(self._k):
index_all = self._clusterAssment[:,0] # 取出样本所属簇的索引值
value = np.nonzero(index_all==i) # 取出所有属于第i个簇的索引值
ptsInClust = data_X[value[0]] # 取出属于第i个簇的所有样本点
self._centroids[i,:] = np.mean(ptsInClust, axis=0) # 计算均值
self._labels = self._clusterAssment[:,0]
self._sse = sum(self._clusterAssment[:,1])
def predict(self, X):
"""根据聚类结果,预测新输入数据所属的簇"""
if not isinstance(X,np.ndarray):
X = np.asarray(X)
m = X.shape[0] # m代表样本数量
preds = np.empty((m,))
# 将每个样本点分配到离它最近的质心所属的簇
for i in range(m):
minDist = np.inf
for j in range(self._k):
distJI = self._calEDist(self._centroids[j,:], X[i,:])
if distJI < minDist:
minDist = distJI
preds[i] = j
return preds
test.py模块:
此处的Iris数据集也可以通过调用sklearn引入
import pandas as pd
import numpy as np
from Kmeans import KMeansClassifier
import matplotlib.pyplot as plt
if __name__=="__main__":
data_X = pd.read_csv(r"iris.csv")
data_X = data_X.drop(data_X.columns[4], axis=1)
data_X = np.array(data_X)
# print(data_X)
# k = 2
k = 3
clf = KMeansClassifier(k) # 实例化KMeansClassifier类
clf.fit(data_X) # 训练模型
cents = clf._centroids
labels = clf._labels
sse = clf._sse
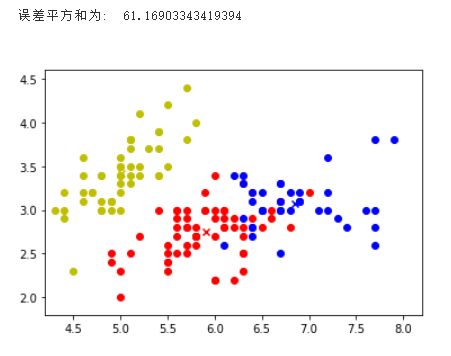
colors = ['r','y','b']
for i in range(k):
index = np.nonzero(labels==i)[0]
# print(index)
x0 = data_X[index, 0]
x1 = data_X[index, 1]
y_i = i
for j in range(len(x0)):
plt.scatter(x0[j], x1[j], color=colors[i])
plt.scatter(cents[i,0], cents[i,1], marker='x', color=colors[i], linewidths=7)
# plt.title("SSE={:.2f}".format(sse))
print("误差平方和为: ", sse)
plt.axis([4.2,8.2,1.8,4.6])
outname = "./Kmeans" + ".png"
plt.savefig(outname)
plt.show()
Output:
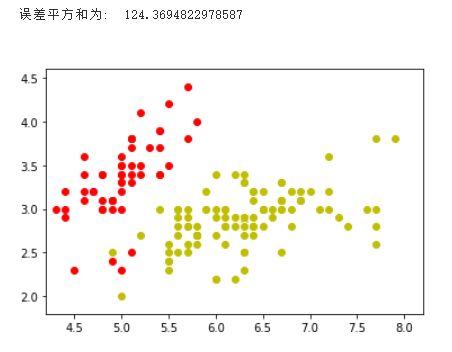
当K选取为2时:
参考资料:《机器学习实战》