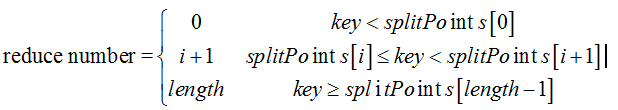修改 Hadoop TeraSort算法 —— 按照LongWritable类型的Key排序
近日,需要用ParMetis对大图数据进行分区,其输入是无向图(邻接表形式)且按照顶点ID排序,于是想到用Hadoop中的TeraSort算法对无向图进行排序。但Hadoop自带TeraSort算法是按照每行数据的前两个字符排序的,不能满足要求。
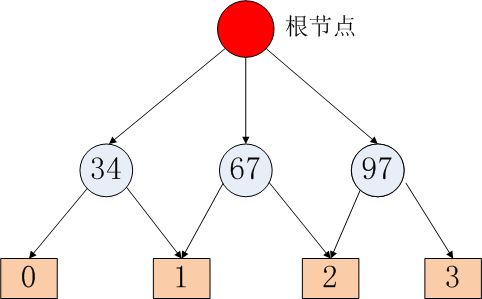
由于图一般都是用邻接表的形式存储,改进的TeraSort算法就是按照顶点ID进行排序,支持有向图和无向图,边上可附加权值。下面以无向图为例讲述数据的输入格式。对于下图,
其输入数据格式如下,以 \t 间隔,每行第一列为顶点ID。
1 2 3 4
3 1 2
2 1 3 4
4 1 2
扩展:只要每行的数据格式满足:key+“\t”+value,其中key为int或long型,value类型任意。修改的TeraSort算法就能按照key来对每一行进行排序。
修改方法:
1. 由于输入格式变化,故首先修改TeraRecordReader类,主要是 boolean next(LongWritable key, Text value)方法,修改如何解析每行数据。代码如下:
public boolean next(LongWritable key, Text value) throws IOException {
if (in.next(junk, line)) {
String[] temp=line.toString().split("\t");
key.set(Long.parseLong(temp[0]));
if(temp.length!=1) {
value.set(line.toString().substring(temp[0].length()+1));
} else {
value.set("");
}
return true;
} else {
return false;
}
}3. 在原TeraSort算法中,每个map task首先从分布式缓存中读取分割点,然后根据分割点简历2-Trie树。map task从split中依次读入每条数据,通过Trie树查找每条记录所对应的reduce task编号。
现在由于是Long型,则不需要构建Trie树。已知分割点是存储在splitPoints[]数组中,按照如下公式计算reduce number,其中length等于splitPoints.length
假设reduce task数目为4(由用户设置),分割点为34、67、97。则分割点和reduce task编号的映射关系如下:
可以看到小于34的对应第0个reduce task,34和67之间的对应第一个reduce task,67和97之间的key对应第2个reduce task,大于等于97的则对应于第3个reduce task。
主要修改int getPartition(LongWritable key, Text value, int numPartitions)方法,如下:
@Override
public int getPartition(LongWritable key, Text value, int numPartitions) {
if(key.get()=splitPoints[i].get() && key.get()
job.setOutputFormat(TextOutputFormat.class);5. 打成Jar包(TeraSort.jar),在集群上运行即可。例如:hadoop jar TeraSort.jar TeraSortTest output 4
注意输入参数为:
6. 上述排好序的文件,依次存在output文件夹下的:part-00000、part-00001、part-00002、part-00003。
使用 hadoop fs -getmerge output output-total 命令后,所有数据都会有序汇总到output-total文件中。
getmerge会按照part-00000、part-00001、part-00002、part-00003的顺序依次把每个文件输出到output-total文件中。代码如下:
/** Copy all files in a directory to one output file (merge). */
public static boolean copyMerge(FileSystem srcFS, Path srcDir,
FileSystem dstFS, Path dstFile,
boolean deleteSource,
Configuration conf, String addString) throws IOException {
dstFile = checkDest(srcDir.getName(), dstFS, dstFile, false);
if (!srcFS.getFileStatus(srcDir).isDir())
return false;
OutputStream out = dstFS.create(dstFile);
try {
FileStatus contents[] = srcFS.listStatus(srcDir);
for (int i = 0; i < contents.length; i++) {
if (!contents[i].isDir()) {
InputStream in = srcFS.open(contents[i].getPath());
try {
IOUtils.copyBytes(in, out, conf, false);
if (addString!=null)
out.write(addString.getBytes("UTF-8"));
} finally {
in.close();
}
}
}
} finally {
out.close();
}part-00000、part-00001、part-00002、part-00003的访问顺序是由namenode获取的,对应其inode节点。
1. TeraInputFormat.java
/**
* Licensed to the Apache Software Foundation (ASF) under one
* or more contributor license agreements. See the NOTICE file
* distributed with this work for additional information
* regarding copyright ownership. The ASF licenses this file
* to you under the Apache License, Version 2.0 (the
* "License"); you may not use this file except in compliance
* with the License. You may obtain a copy of the License at
*
* http://www.apache.org/licenses/LICENSE-2.0
*
* Unless required by applicable law or agreed to in writing, software
* distributed under the License is distributed on an "AS IS" BASIS,
* WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
* See the License for the specific language governing permissions and
* limitations under the License.
*/
package com.undirected.graph.sort;
import java.io.IOException;
import java.util.ArrayList;
import org.apache.hadoop.conf.Configuration;
import org.apache.hadoop.fs.FileSystem;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.NullWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapred.FileInputFormat;
import org.apache.hadoop.mapred.FileSplit;
import org.apache.hadoop.mapred.InputSplit;
import org.apache.hadoop.mapred.JobConf;
import org.apache.hadoop.mapred.LineRecordReader;
import org.apache.hadoop.mapred.RecordReader;
import org.apache.hadoop.mapred.Reporter;
import org.apache.hadoop.util.IndexedSortable;
import org.apache.hadoop.util.QuickSort;
/**
* An input format that reads the first 10 characters of each line as the key
* and the rest of the line as the value. Both key and value are represented
* as Text.
*/
public class TeraInputFormat extends FileInputFormat {
static final String PARTITION_FILENAME = "_partition.lst";
static final String SAMPLE_SIZE = "terasort.partitions.sample";
private static JobConf lastConf = null;
private static InputSplit[] lastResult = null;
static class TextSampler implements IndexedSortable {
private ArrayList records = new ArrayList();
public int compare(int i, int j) {
LongWritable left = records.get(i);
LongWritable right = records.get(j);
return left.compareTo(right);
}
public void swap(int i, int j) {
LongWritable left = records.get(i);
LongWritable right = records.get(j);
records.set(j, left);
records.set(i, right);
}
public void addKey(LongWritable key) {
records.add(key);
}
/**
* Find the split points for a given sample. The sample keys are sorted
* and down sampled to find even split points for the partitions. The
* returned keys should be the start of their respective partitions.
* @param numPartitions the desired number of partitions
* @return an array of size numPartitions - 1 that holds the split points
*/
LongWritable[] createPartitions(int numPartitions) {
int numRecords = records.size();
System.out.println("Making " + numPartitions + " from " + numRecords +
" records");
if (numPartitions > numRecords) {
throw new IllegalArgumentException
("Requested more partitions than input keys (" + numPartitions +
" > " + numRecords + ")");
}
new QuickSort().sort(this, 0, records.size());
float stepSize = numRecords / (float) numPartitions;
System.out.println("Step size is " + stepSize);
LongWritable[] result = new LongWritable[numPartitions-1];
for(int i=1; i < numPartitions; ++i) {
result[i-1] = records.get(Math.round(stepSize * i));
}
// System.out.println("result :"+Arrays.toString(result));
return result;
}
}
/**
* Use the input splits to take samples of the input and generate sample
* keys. By default reads 100,000 keys from 10 locations in the input, sorts
* them and picks N-1 keys to generate N equally sized partitions.
* @param conf the job to sample
* @param partFile where to write the output file to
* @throws IOException if something goes wrong
*/
public static void writePartitionFile(JobConf conf,
Path partFile) throws IOException {
TeraInputFormat inFormat = new TeraInputFormat();
TextSampler sampler = new TextSampler();
LongWritable key = new LongWritable();
Text value = new Text();
int partitions = conf.getNumReduceTasks();
long sampleSize = conf.getLong(SAMPLE_SIZE, 100000);
InputSplit[] splits = inFormat.getSplits(conf, conf.getNumMapTasks());
int samples = Math.min(10, splits.length);
long recordsPerSample = sampleSize / samples;
int sampleStep = splits.length / samples;
long records = 0;
// take N samples from different parts of the input
for(int i=0; i < samples; ++i) {
RecordReader reader =
inFormat.getRecordReader(splits[sampleStep * i], conf, null);
while (reader.next(key, value)) {
sampler.addKey(key);
key=new LongWritable();
records += 1;
if ((i+1) * recordsPerSample <= records) {
break;
}
}
}
FileSystem outFs = partFile.getFileSystem(conf);
if (outFs.exists(partFile)) {
outFs.delete(partFile, false);
}
SequenceFile.Writer writer =
SequenceFile.createWriter(outFs, conf, partFile, LongWritable.class,
NullWritable.class);
NullWritable nullValue = NullWritable.get();
for(LongWritable split : sampler.createPartitions(partitions)) {
writer.append(split, nullValue);
}
writer.close();
}
static class TeraRecordReader implements RecordReader {
private LineRecordReader in;
private LongWritable junk = new LongWritable();
private Text line = new Text();
public TeraRecordReader(Configuration job,
FileSplit split) throws IOException {
in = new LineRecordReader(job, split);
}
public void close() throws IOException {
in.close();
}
public LongWritable createKey() {
return new LongWritable();
}
public Text createValue() {
return new Text();
}
public long getPos() throws IOException {
return in.getPos();
}
public float getProgress() throws IOException {
return in.getProgress();
}
public boolean next(LongWritable key, Text value) throws IOException {
if (in.next(junk, line)) {
String[] temp=line.toString().split("\t");
key.set(Long.parseLong(temp[0]));
if(temp.length!=1) {
value.set(line.toString().substring(temp[0].length()+1));
} else {
value.set("");
}
return true;
} else {
return false;
}
}
}
@Override
public RecordReader
getRecordReader(InputSplit split,
JobConf job,
Reporter reporter) throws IOException {
return new TeraRecordReader(job, (FileSplit) split);
}
@Override
public InputSplit[] getSplits(JobConf conf, int splits) throws IOException {
if (conf == lastConf) {
return lastResult;
}
lastConf = conf;
lastResult = super.getSplits(conf, splits);
return lastResult;
}
}
2. TeraSort.java
package com.undirected.graph.sort;
import java.io.IOException;
import java.net.URI;
import java.util.ArrayList;
import java.util.List;
import org.apache.commons.logging.Log;
import org.apache.commons.logging.LogFactory;
import org.apache.hadoop.conf.Configured;
import org.apache.hadoop.filecache.DistributedCache;
import org.apache.hadoop.fs.FileSystem;
import org.apache.hadoop.fs.Path;
import org.apache.hadoop.io.LongWritable;
import org.apache.hadoop.io.NullWritable;
import org.apache.hadoop.io.SequenceFile;
import org.apache.hadoop.io.Text;
import org.apache.hadoop.mapred.FileOutputFormat;
import org.apache.hadoop.mapred.JobClient;
import org.apache.hadoop.mapred.JobConf;
import org.apache.hadoop.mapred.Partitioner;
import org.apache.hadoop.mapred.TextOutputFormat;
import org.apache.hadoop.util.Tool;
import org.apache.hadoop.util.ToolRunner;
public class TeraSort extends Configured implements Tool{
private static final Log LOG = LogFactory.getLog(TeraSort.class);
/**
* A partitioner that splits text keys into roughly equal partitions
* in a global sorted order.
*/
static class TotalOrderPartitioner implements Partitioner{
private LongWritable[] splitPoints;
/**
* Read the cut points from the given sequence file.
* @param fs the file system
* @param p the path to read
* @param job the job config
* @return the strings to split the partitions on
* @throws IOException
*/
private static LongWritable[] readPartitions(FileSystem fs, Path p,
JobConf job) throws IOException {
SequenceFile.Reader reader = new SequenceFile.Reader(fs, p, job);
List parts = new ArrayList();
LongWritable key = new LongWritable();
NullWritable value = NullWritable.get();
while (reader.next(key, value)) {
parts.add(key);
key = new LongWritable();
}
reader.close();
return parts.toArray(new LongWritable[parts.size()]);
}
@Override
public void configure(JobConf job) {
try {
FileSystem fs = FileSystem.getLocal(job);
Path partFile = new Path(TeraInputFormat.PARTITION_FILENAME);
splitPoints = readPartitions(fs, partFile, job);
} catch (IOException ie) {
throw new IllegalArgumentException("can't read paritions file", ie);
}
}
@Override
public int getPartition(LongWritable key, Text value, int numPartitions) {
if(key.get()=splitPoints[i].get() && key.get()