提高神经网络的学习方式Improving the way neural networks learn-2
Exercise
- As discussed above, one way of expanding the MNIST training data is to use small rotations of training images. What's a problem that might occur if we allow arbitrarily large rotations of training images?
An aside on big data and what it means to compare classification accuracies: Let's look again at how our neural network's accuracy varies with training set size:
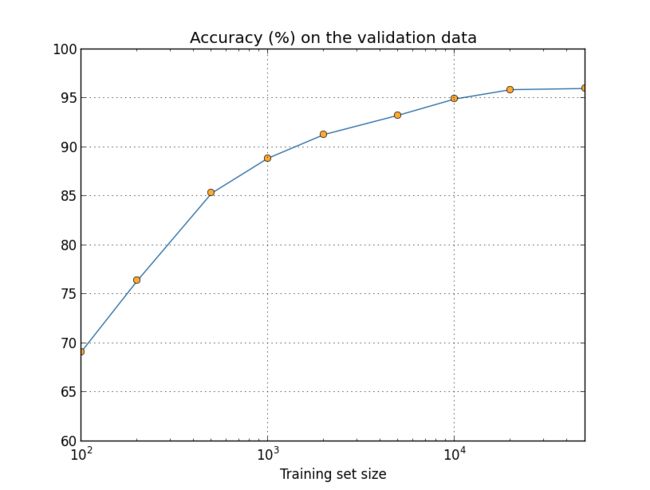
Suppose that instead of using a neural network we use some other machine learning technique to classify digits. For instance, let's try using the support vector machines (SVM) which we met briefly back in Chapter 1. As was the case in Chapter 1, don't worry if you're not familiar with SVMs, we don't need to understand their details. Instead, we'll use the SVM supplied by the scikit-learn library. Here's how SVM performance varies as a function of training set size. I've plotted the neural net results as well, to make comparison easy**This graph was produced with the programmore_data.py (as were the last few graphs).:
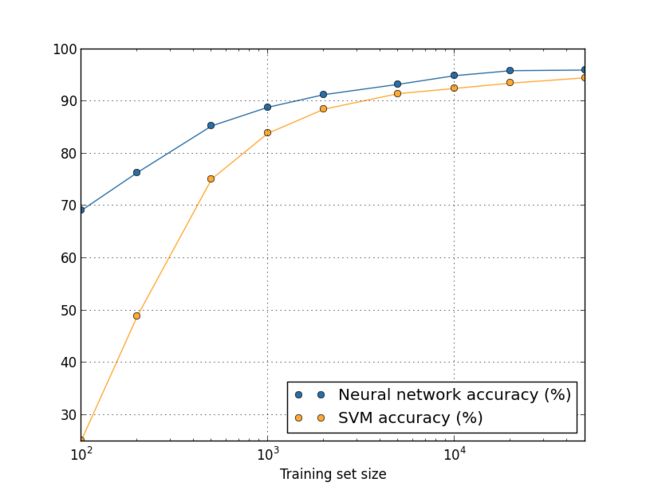
Probably the first thing that strikes you about this graph is that our neural network outperforms the SVM for every training set size. That's nice, although you shouldn't read too much into it, since I just used the out-of-the-box settings from scikit-learn's SVM, while we've done a fair bit of work improving our neural network. A more subtle but more interesting fact about the graph is that if we train our SVM using 50,000 images then it actually has better performance (94.48 percent accuracy) than our neural network does when trained using 5,000 images (93.24 percent accuracy). In other words, more training data can sometimes compensate for differences in the machine learning algorithm used.
Something even more interesting can occur. Suppose we're trying to solve a problem using two machine learning algorithms, algorithm A and algorithm B. It sometimes happens that algorithm A will outperform algorithm B with one set of training data, while algorithm B will outperform algorithm A with a different set of training data. We don't see that above - it would require the two graphs to cross - but it does happen**Striking examples may be found in Scaling to very very large corpora for natural language disambiguation, by Michele Banko and Eric Brill (2001).. The correct response to the question "Is algorithm A better than algorithm B?" is really: "What training data set are you using?"
All this is a caution to keep in mind, both when doing development, and when reading research papers. Many papers focus on finding new tricks to wring out improved performance on standard benchmark data sets. "Our whiz-bang technique gave us an improvement of X percent on standard benchmark Y" is a canonical form of research claim. Such claims are often genuinely interesting, but they must be understood as applying only in the context of the specific training data set used. Imagine an alternate history in which the people who originally created the benchmark data set had a larger research grant. They might have used the extra money to collect more training data. It's entirely possible that the "improvement" due to the whiz-bang technique would disappear on a larger data set. In other words, the purported improvement might be just an accident of history. The message to take away, especially in practical applications, is that what we want is both better algorithms and better training data. It's fine to look for better algorithms, but make sure you're not focusing on better algorithms to the exclusion of easy wins getting more or better training data.
Problem
- (Research problem) How do our machine learning algorithms perform in the limit of very large data sets? For any given algorithm it's natural to attempt to define a notion of asymptotic performance in the limit of truly big data. A quick-and-dirty approach to this problem is to simply try fitting curves to graphs like those shown above, and then to extrapolate the fitted curves out to infinity. An objection to this approach is that different approaches to curve fitting will give different notions of asymptotic performance. Can you find a principled justification for fitting to some particular class of curves? If so, compare the asymptotic performance of several different machine learning algorithms.
Summing up: We've now completed our dive into overfitting and regularization. Of course, we'll return again to the issue. As I've mentioned several times, overfitting is a major problem in neural networks, especially as computers get more powerful, and we have the ability to train larger networks. As a result there's a pressing need to develop powerful regularization techniques to reduce overfitting, and this is an extremely active area of current work.
Weight initialization
When we create our neural networks, we have to make choices for the initial weights and biases. Up to now, we've been choosing them according to a prescription which I discussed only briefly back in Chapter 1. Just to remind you, that prescription was to choose both the weights and biases using independent Gaussian random variables, normalized to have mean 0 0 and standard deviation 1 1. While this approach has worked well, it was quite ad hoc, and it's worth revisiting to see if we can find a better way of setting our initial weights and biases, and perhaps help our neural networks learn faster.
It turns out that we can do quite a bit better than initializing with normalized Gaussians. To see why, suppose we're working with a network with a large number - say 1,000 1,000 - of input neurons. And let's suppose we've used normalized Gaussians to initialize the weights connecting to the first hidden layer. For now I'm going to concentrate specifically on the weights connecting the input neurons to the first neuron in the hidden layer, and ignore the rest of the network:

We'll suppose for simplicity that we're trying to train using a training input x x in which half the input neurons are on, i.e., set to 1 1, and half the input neurons are off, i.e., set to 0 0. The argument which follows applies more generally, but you'll get the gist from this special case. Let's consider the weighted sum z=∑jwjxj+b z=∑jwjxj+b of inputs to our hidden neuron. 500 500 terms in this sum vanish, because the corresponding input xj xj is zero. And so z z is a sum over a total of 501 501 normalized Gaussian random variables, accounting for the 500 500weight terms and the 1 1 extra bias term. Thus z z is itself distributed as a Gaussian with mean zero and standard deviation 501−−−√≈22.4 501≈22.4. That is, z z has a very broad Gaussian distribution, not sharply peaked at all:
In particular, we can see from this graph that it's quite likely that |z| |z|will be pretty large, i.e., either z≫1 z≫1 or z≪−1 z≪−1. If that's the case then the output σ(z) σ(z) from the hidden neuron will be very close to either 1 1 or 0 0. That means our hidden neuron will have saturated. And when that happens, as we know, making small changes in the weights will make only absolutely miniscule changes in the activation of our hidden neuron. That miniscule change in the activation of the hidden neuron will, in turn, barely affect the rest of the neurons in the network at all, and we'll see a correspondingly miniscule change in the cost function. As a result, those weights will only learn very slowly when we use the gradient descent algorithm**We discussed this in more detail in Chapter 2, where we used the equations of backpropagationto show that weights input to saturated neurons learned slowly.. It's similar to the problem we discussed earlier in this chapter, in which output neurons which saturated on the wrong value caused learning to slow down. We addressed that earlier problem with a clever choice of cost function. Unfortunately, while that helped with saturated output neurons, it does nothing at all for the problem with saturated hidden neurons.
I've been talking about the weights input to the first hidden layer. Of course, similar arguments apply also to later hidden layers: if the weights in later hidden layers are initialized using normalized Gaussians, then activations will often be very close to 0 0 or 1 1, and learning will proceed very slowly.
Is there some way we can choose better initializations for the weights and biases, so that we don't get this kind of saturation, and so avoid a learning slowdown? Suppose we have a neuron with nin nininput weights. Then we shall initialize those weights as Gaussian random variables with mean 0 0 and standard deviation 1/nin−−−√ 1/nin. That is, we'll squash the Gaussians down, making it less likely that our neuron will saturate. We'll continue to choose the bias as a Gaussian with mean 0 0 and standard deviation 1 1, for reasons I'll return to in a moment. With these choices, the weighted sum z=∑jwjxj+b z=∑jwjxj+b will again be a Gaussian random variable with mean 0 0, but it'll be much more sharply peaked than it was before. Suppose, as we did earlier, that 500 500 of the inputs are zero and 500 500are 1 1. Then it's easy to show (see the exercise below) that z z has a Gaussian distribution with mean 0 0 and standard deviation 3/2−−−√=1.22… 3/2=1.22…. This is much more sharply peaked than before, so much so that even the graph below understates the situation, since I've had to rescale the vertical axis, when compared to the earlier graph:
Such a neuron is much less likely to saturate, and correspondingly much less likely to have problems with a learning slowdown.
Exercise
- Verify that the standard deviation of z=∑jwjxj+b z=∑jwjxj+b in the paragraph above is 3/2−−−√ 3/2. It may help to know that: (a) the variance of a sum of independent random variables is the sum of the variances of the individual random variables; and (b) the variance is the square of the standard deviation.
I stated above that we'll continue to initialize the biases as before, as Gaussian random variables with a mean of 0 0 and a standard deviation of 1 1. This is okay, because it doesn't make it too much more likely that our neurons will saturate. In fact, it doesn't much matter how we initialize the biases, provided we avoid the problem with saturation. Some people go so far as to initialize all the biases to 0 0, and rely on gradient descent to learn appropriate biases. But since it's unlikely to make much difference, we'll continue with the same initialization procedure as before.
Let's compare the results for both our old and new approaches to weight initialization, using the MNIST digit classification task. As before, we'll use 30 30 hidden neurons, a mini-batch size of 10 10, a regularization parameter λ=5.0 λ=5.0, and the cross-entropy cost function. We will decrease the learning rate slightly from η=0.5 η=0.5 to 0.1 0.1, since that makes the results a little more easily visible in the graphs. We can train using the old method of weight initialization:
>>> import mnist_loader
>>> training_data, validation_data, test_data = \
... mnist_loader.load_data_wrapper()
>>> import network2
>>> net = network2.Network([784, 30, 10], cost=network2.CrossEntropyCost)
>>> net.large_weight_initializer()
>>> net.SGD(training_data, 30, 10, 0.1, lmbda = 5.0,
... evaluation_data=validation_data,
... monitor_evaluation_accuracy=True)
>>> net = network2.Network([784, 30, 10], cost=network2.CrossEntropyCost)
>>> net.SGD(training_data, 30, 10, 0.1, lmbda = 5.0,
... evaluation_data=validation_data,
... monitor_evaluation_accuracy=True)
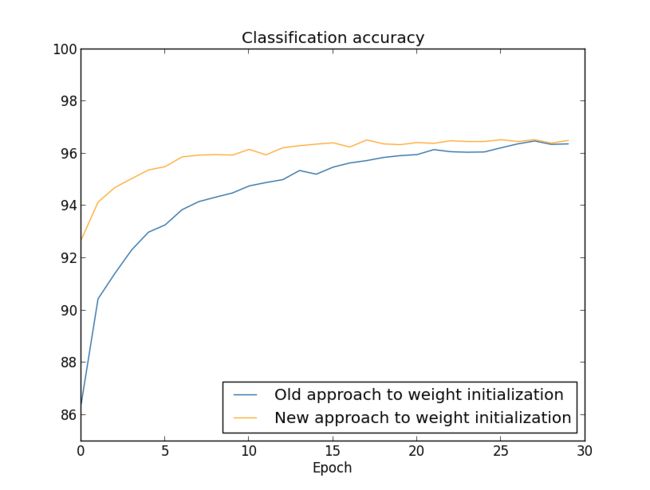
In both cases, we end up with a classification accuracy somewhat over 96 percent. The final classification accuracy is almost exactly the same in the two cases. But the new initialization technique brings us there much, much faster. At the end of the first epoch of training the old approach to weight initialization has a classification accuracy under 87 percent, while the new approach is already almost 93 percent. What appears to be going on is that our new approach to weight initialization starts us off in a much better regime, which lets us get good results much more quickly. The same phenomenon is also seen if we plot results with 100 100 hidden neurons:
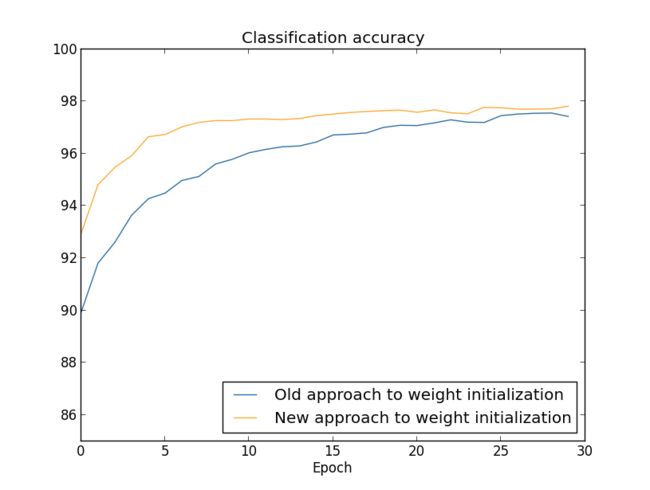
In this case, the two curves don't quite meet. However, my experiments suggest that with just a few more epochs of training (not shown) the accuracies become almost exactly the same. So on the basis of these experiments it looks as though the improved weight initialization only speeds up learning, it doesn't change the final performance of our networks. However, in Chapter 4 we'll see examples of neural networks where the long-run behaviour is significantly better with the 1/nin−−−√ 1/nin weight initialization. Thus it's not only the speed of learning which is improved, it's sometimes also the final performance.
The 1/nin−−−√ 1/nin approach to weight initialization helps improve the way our neural nets learn. Other techniques for weight initialization have also been proposed, many building on this basic idea. I won't review the other approaches here, since 1/nin−−−√ 1/nin works well enough for our purposes. If you're interested in looking further, I recommend looking at the discussion on pages 14 and 15 of a 2012 paper by Yoshua Bengio**Practical Recommendations for Gradient-Based Training of Deep Architectures, by Yoshua Bengio (2012)., as well as the references therein.
Problem
- Connecting regularization and the improved method of weight initialization L2 regularization sometimes automatically gives us something similar to the new approach to weight initialization. Suppose we are using the old approach to weight initialization. Sketch a heuristic argument that: (1) supposing λ λ is not too small, the first epochs of training will be dominated almost entirely by weight decay; (2) provided ηλ≪n ηλ≪n the weights will decay by a factor of exp(−ηλ/m) exp(−ηλ/m) per epoch; and (3) supposing λ λ is not too large, the weight decay will tail off when the weights are down to a size around 1/n√ 1/n, where n n is the total number of weights in the network. Argue that these conditions are all satisfied in the examples graphed in this section.
Handwriting recognition revisited: the code
Let's implement the ideas we've discussed in this chapter. We'll develop a new program, network2.py, which is an improved version of the program network.py we developed in Chapter 1. If you haven't looked at network.py in a while then you may find it helpful to spend a few minutes quickly reading over the earlier discussion. It's only 74 lines of code, and is easily understood.
As was the case in network.py, the star of network2.py is the Networkclass, which we use to represent our neural networks. We initialize an instance of Network with a list of sizes for the respective layers in the network, and a choice for the cost to use, defaulting to the cross-entropy:
class Network(object):
def __init__(self, sizes, cost=CrossEntropyCost):
self.num_layers = len(sizes)
self.sizes = sizes
self.default_weight_initializer()
self.cost=cost
The first couple of lines of the __init__ method are the same as innetwork.py, and are pretty self-explanatory. But the next two lines are new, and we need to understand what they're doing in detail.
Let's start by examining the default_weight_initializer method. This makes use of our new and improved approach to weight initialization. As we've seen, in that approach the weights input to a neuron are initialized as Gaussian random variables with mean 0 and standard deviation 1 1 divided by the square root of the number of connections input to the neuron. Also in this method we'll initialize the biases, using Gaussian random variables with mean 0 0and standard deviation 1 1. Here's the code:
def default_weight_initializer(self):
self.biases = [np.random.randn(y, 1) for y in self.sizes[1:]]
self.weights = [np.random.randn(y, x)/np.sqrt(x)
for x, y in zip(self.sizes[:-1], self.sizes[1:])]
To understand the code, it may help to recall that np is the Numpy library for doing linear algebra. We'll import Numpy at the beginning of our program. Also, notice that we don't initialize any biases for the first layer of neurons. We avoid doing this because the first layer is an input layer, and so any biases would not be used. We did exactly the same thing in network.py.
Complementing the default_weight_initializer we'll also include alarge_weight_initializer method. This method initializes the weights and biases using the old approach from Chapter 1, with both weights and biases initialized as Gaussian random variables with mean 0 0 and standard deviation 1 1. The code is, of course, only a tiny bit different from the default_weight_initializer:
def large_weight_initializer(self):
self.biases = [np.random.randn(y, 1) for y in self.sizes[1:]]
self.weights = [np.random.randn(y, x)
for x, y in zip(self.sizes[:-1], self.sizes[1:])]
I've included the large_weight_initializer method mostly as a convenience to make it easier to compare the results in this chapter to those in Chapter 1. I can't think of many practical situations where I would recommend using it!
The second new thing in Network's __init__ method is that we now initialize a cost attribute. To understand how that works, let's look at the class we use to represent the cross-entropy cost**If you're not familiar with Python's static methods you can ignore the @staticmethoddecorators, and just treat fn and delta as ordinary methods. If you're curious about details, all @staticmethod does is tell the Python interpreter that the method which follows doesn't depend on the object in any way. That's why self isn't passed as a parameter to the fnand delta methods.:
class CrossEntropyCost(object):
@staticmethod
def fn(a, y):
return np.sum(np.nan_to_num(-y*np.log(a)-(1-y)*np.log(1-a)))
@staticmethod
def delta(z, a, y):
return (a-y)
Let's break this down. The first thing to observe is that even though the cross-entropy is, mathematically speaking, a function, we've implemented it as a Python class, not a Python function. Why have I made that choice? The reason is that the cost plays two different roles in our network. The obvious role is that it's a measure of how well an output activation, a, matches the desired output, y. This role is captured by the CrossEntropyCost.fn method. (Note, by the way, that the np.nan_to_num call inside CrossEntropyCost.fn ensures that Numpy deals correctly with the log of numbers very close to zero.) But there's also a second way the cost function enters our network. Recall from Chapter 2 that when running the backpropagation algorithm we need to compute the network's output error, δL δL. The form of the output error depends on the choice of cost function: different cost function, different form for the output error. For the cross-entropy the output error is, as we saw in Equation (66),
In a similar way, network2.py also contains a class to represent the quadratic cost function. This is included for comparison with the results of Chapter 1, since going forward we'll mostly use the cross entropy. The code is just below. The QuadraticCost.fn method is a straightforward computation of the quadratic cost associated to the actual output, a, and the desired output, y. The value returned byQuadraticCost.delta is based on the expression (30) for the output error for the quadratic cost, which we derived back in Chapter 2.
class QuadraticCost(object):
@staticmethod
def fn(a, y):
return 0.5*np.linalg.norm(a-y)**2
@staticmethod
def delta(z, a, y):
return (a-y) * sigmoid_prime(z)
We've now understood the main differences between network2.py andnetwork.py. It's all pretty simple stuff. There are a number of smaller changes, which I'll discuss below, including the implementation of L2 regularization. Before getting to that, let's look at the complete code for network2.py. You don't need to read all the code in detail, but it is worth understanding the broad structure, and in particular reading the documentation strings, so you understand what each piece of the program is doing. Of course, you're also welcome to delve as deeply as you wish! If you get lost, you may wish to continue reading the prose below, and return to the code later. Anyway, here's the code:
"""network2.py
~~~~~~~~~~~~~~
An improved version of network.py, implementing the stochastic
gradient descent learning algorithm for a feedforward neural network.
Improvements include the addition of the cross-entropy cost function,
regularization, and better initialization of network weights. Note
that I have focused on making the code simple, easily readable, and
easily modifiable. It is not optimized, and omits many desirable
features.
"""
#### Libraries
# Standard library
import json
import random
import sys
# Third-party libraries
import numpy as np
#### Define the quadratic and cross-entropy cost functions
class QuadraticCost(object):
@staticmethod
def fn(a, y):
"""Return the cost associated with an output ``a`` and desired output
``y``.
"""
return 0.5*np.linalg.norm(a-y)**2
@staticmethod
def delta(z, a, y):
"""Return the error delta from the output layer."""
return (a-y) * sigmoid_prime(z)
class CrossEntropyCost(object):
@staticmethod
def fn(a, y):
"""Return the cost associated with an output ``a`` and desired output
``y``. Note that np.nan_to_num is used to ensure numerical
stability. In particular, if both ``a`` and ``y`` have a 1.0
in the same slot, then the expression (1-y)*np.log(1-a)
returns nan. The np.nan_to_num ensures that that is converted
to the correct value (0.0).
"""
return np.sum(np.nan_to_num(-y*np.log(a)-(1-y)*np.log(1-a)))
@staticmethod
def delta(z, a, y):
"""Return the error delta from the output layer. Note that the
parameter ``z`` is not used by the method. It is included in
the method's parameters in order to make the interface
consistent with the delta method for other cost classes.
"""
return (a-y)
#### Main Network class
class Network(object):
def __init__(self, sizes, cost=CrossEntropyCost):
"""The list ``sizes`` contains the number of neurons in the respective
layers of the network. For example, if the list was [2, 3, 1]
then it would be a three-layer network, with the first layer
containing 2 neurons, the second layer 3 neurons, and the
third layer 1 neuron. The biases and weights for the network
are initialized randomly, using
``self.default_weight_initializer`` (see docstring for that
method).
"""
self.num_layers = len(sizes)
self.sizes = sizes
self.default_weight_initializer()
self.cost=cost
def default_weight_initializer(self):
"""Initialize each weight using a Gaussian distribution with mean 0
and standard deviation 1 over the square root of the number of
weights connecting to the same neuron. Initialize the biases
using a Gaussian distribution with mean 0 and standard
deviation 1.
Note that the first layer is assumed to be an input layer, and
by convention we won't set any biases for those neurons, since
biases are only ever used in computing the outputs from later
layers.
"""
self.biases = [np.random.randn(y, 1) for y in self.sizes[1:]]
self.weights = [np.random.randn(y, x)/np.sqrt(x)
for x, y in zip(self.sizes[:-1], self.sizes[1:])]
def large_weight_initializer(self):
"""Initialize the weights using a Gaussian distribution with mean 0
and standard deviation 1. Initialize the biases using a
Gaussian distribution with mean 0 and standard deviation 1.
Note that the first layer is assumed to be an input layer, and
by convention we won't set any biases for those neurons, since
biases are only ever used in computing the outputs from later
layers.
This weight and bias initializer uses the same approach as in
Chapter 1, and is included for purposes of comparison. It
will usually be better to use the default weight initializer
instead.
"""
self.biases = [np.random.randn(y, 1) for y in self.sizes[1:]]
self.weights = [np.random.randn(y, x)
for x, y in zip(self.sizes[:-1], self.sizes[1:])]
def feedforward(self, a):
"""Return the output of the network if ``a`` is input."""
for b, w in zip(self.biases, self.weights):
a = sigmoid(np.dot(w, a)+b)
return a
def SGD(self, training_data, epochs, mini_batch_size, eta,
lmbda = 0.0,
evaluation_data=None,
monitor_evaluation_cost=False,
monitor_evaluation_accuracy=False,
monitor_training_cost=False,
monitor_training_accuracy=False):
"""Train the neural network using mini-batch stochastic gradient
descent. The ``training_data`` is a list of tuples ``(x, y)``
representing the training inputs and the desired outputs. The
other non-optional parameters are self-explanatory, as is the
regularization parameter ``lmbda``. The method also accepts
``evaluation_data``, usually either the validation or test
data. We can monitor the cost and accuracy on either the
evaluation data or the training data, by setting the
appropriate flags. The method returns a tuple containing four
lists: the (per-epoch) costs on the evaluation data, the
accuracies on the evaluation data, the costs on the training
data, and the accuracies on the training data. All values are
evaluated at the end of each training epoch. So, for example,
if we train for 30 epochs, then the first element of the tuple
will be a 30-element list containing the cost on the
evaluation data at the end of each epoch. Note that the lists
are empty if the corresponding flag is not set.
"""
if evaluation_data: n_data = len(evaluation_data)
n = len(training_data)
evaluation_cost, evaluation_accuracy = [], []
training_cost, training_accuracy = [], []
for j in xrange(epochs):
random.shuffle(training_data)
mini_batches = [
training_data[k:k+mini_batch_size]
for k in xrange(0, n, mini_batch_size)]
for mini_batch in mini_batches:
self.update_mini_batch(
mini_batch, eta, lmbda, len(training_data))
print "Epoch %s training complete" % j
if monitor_training_cost:
cost = self.total_cost(training_data, lmbda)
training_cost.append(cost)
print "Cost on training data: {}".format(cost)
if monitor_training_accuracy:
accuracy = self.accuracy(training_data, convert=True)
training_accuracy.append(accuracy)
print "Accuracy on training data: {} / {}".format(
accuracy, n)
if monitor_evaluation_cost:
cost = self.total_cost(evaluation_data, lmbda, convert=True)
evaluation_cost.append(cost)
print "Cost on evaluation data: {}".format(cost)
if monitor_evaluation_accuracy:
accuracy = self.accuracy(evaluation_data)
evaluation_accuracy.append(accuracy)
print "Accuracy on evaluation data: {} / {}".format(
self.accuracy(evaluation_data), n_data)
print
return evaluation_cost, evaluation_accuracy, \
training_cost, training_accuracy
def update_mini_batch(self, mini_batch, eta, lmbda, n):
"""Update the network's weights and biases by applying gradient
descent using backpropagation to a single mini batch. The
``mini_batch`` is a list of tuples ``(x, y)``, ``eta`` is the
learning rate, ``lmbda`` is the regularization parameter, and
``n`` is the total size of the training data set.
"""
nabla_b = [np.zeros(b.shape) for b in self.biases]
nabla_w = [np.zeros(w.shape) for w in self.weights]
for x, y in mini_batch:
delta_nabla_b, delta_nabla_w = self.backprop(x, y)
nabla_b = [nb+dnb for nb, dnb in zip(nabla_b, delta_nabla_b)]
nabla_w = [nw+dnw for nw, dnw in zip(nabla_w, delta_nabla_w)]
self.weights = [(1-eta*(lmbda/n))*w-(eta/len(mini_batch))*nw
for w, nw in zip(self.weights, nabla_w)]
self.biases = [b-(eta/len(mini_batch))*nb
for b, nb in zip(self.biases, nabla_b)]
def backprop(self, x, y):
"""Return a tuple ``(nabla_b, nabla_w)`` representing the
gradient for the cost function C_x. ``nabla_b`` and
``nabla_w`` are layer-by-layer lists of numpy arrays, similar
to ``self.biases`` and ``self.weights``."""
nabla_b = [np.zeros(b.shape) for b in self.biases]
nabla_w = [np.zeros(w.shape) for w in self.weights]
# feedforward
activation = x
activations = [x] # list to store all the activations, layer by layer
zs = [] # list to store all the z vectors, layer by layer
for b, w in zip(self.biases, self.weights):
z = np.dot(w, activation)+b
zs.append(z)
activation = sigmoid(z)
activations.append(activation)
# backward pass
delta = (self.cost).delta(zs[-1], activations[-1], y)
nabla_b[-1] = delta
nabla_w[-1] = np.dot(delta, activations[-2].transpose())
# Note that the variable l in the loop below is used a little
# differently to the notation in Chapter 2 of the book. Here,
# l = 1 means the last layer of neurons, l = 2 is the
# second-last layer, and so on. It's a renumbering of the
# scheme in the book, used here to take advantage of the fact
# that Python can use negative indices in lists.
for l in xrange(2, self.num_layers):
z = zs[-l]
sp = sigmoid_prime(z)
delta = np.dot(self.weights[-l+1].transpose(), delta) * sp
nabla_b[-l] = delta
nabla_w[-l] = np.dot(delta, activations[-l-1].transpose())
return (nabla_b, nabla_w)
def accuracy(self, data, convert=False):
"""Return the number of inputs in ``data`` for which the neural
network outputs the correct result. The neural network's
output is assumed to be the index of whichever neuron in the
final layer has the highest activation.
The flag ``convert`` should be set to False if the data set is
validation or test data (the usual case), and to True if the
data set is the training data. The need for this flag arises
due to differences in the way the results ``y`` are
represented in the different data sets. In particular, it
flags whether we need to convert between the different
representations. It may seem strange to use different
representations for the different data sets. Why not use the
same representation for all three data sets? It's done for
efficiency reasons -- the program usually evaluates the cost
on the training data and the accuracy on other data sets.
These are different types of computations, and using different
representations speeds things up. More details on the
representations can be found in
mnist_loader.load_data_wrapper.
"""
if convert:
results = [(np.argmax(self.feedforward(x)), np.argmax(y))
for (x, y) in data]
else:
results = [(np.argmax(self.feedforward(x)), y)
for (x, y) in data]
return sum(int(x == y) for (x, y) in results)
def total_cost(self, data, lmbda, convert=False):
"""Return the total cost for the data set ``data``. The flag
``convert`` should be set to False if the data set is the
training data (the usual case), and to True if the data set is
the validation or test data. See comments on the similar (but
reversed) convention for the ``accuracy`` method, above.
"""
cost = 0.0
for x, y in data:
a = self.feedforward(x)
if convert: y = vectorized_result(y)
cost += self.cost.fn(a, y)/len(data)
cost += 0.5*(lmbda/len(data))*sum(
np.linalg.norm(w)**2 for w in self.weights)
return cost
def save(self, filename):
"""Save the neural network to the file ``filename``."""
data = {"sizes": self.sizes,
"weights": [w.tolist() for w in self.weights],
"biases": [b.tolist() for b in self.biases],
"cost": str(self.cost.__name__)}
f = open(filename, "w")
json.dump(data, f)
f.close()
#### Loading a Network
def load(filename):
"""Load a neural network from the file ``filename``. Returns an
instance of Network.
"""
f = open(filename, "r")
data = json.load(f)
f.close()
cost = getattr(sys.modules[__name__], data["cost"])
net = Network(data["sizes"], cost=cost)
net.weights = [np.array(w) for w in data["weights"]]
net.biases = [np.array(b) for b in data["biases"]]
return net
#### Miscellaneous functions
def vectorized_result(j):
"""Return a 10-dimensional unit vector with a 1.0 in the j'th position
and zeroes elsewhere. This is used to convert a digit (0...9)
into a corresponding desired output from the neural network.
"""
e = np.zeros((10, 1))
e[j] = 1.0
return e
def sigmoid(z):
"""The sigmoid function."""
return 1.0/(1.0+np.exp(-z))
def sigmoid_prime(z):
"""Derivative of the sigmoid function."""
return sigmoid(z)*(1-sigmoid(z))
One of the more interesting changes in the code is to include L2 regularization. Although this is a major conceptual change, it's so trivial to implement that it's easy to miss in the code. For the most part it just involves passing the parameter lmbda to various methods, notably the Network.SGD method. The real work is done in a single line of the program, the fourth-last line of the Network.update_mini_batchmethod. That's where we modify the gradient descent update rule to include weight decay. But although the modification is tiny, it has a big impact on results!
This is, by the way, common when implementing new techniques in neural networks. We've spent thousands of words discussing regularization. It's conceptually quite subtle and difficult to understand. And yet it was trivial to add to our program! It occurs surprisingly often that sophisticated techniques can be implemented with small changes to code.
Another small but important change to our code is the addition of several optional flags to the stochastic gradient descent method,Network.SGD. These flags make it possible to monitor the cost and accuracy either on the training_data or on a set of evaluation_datawhich can be passed to Network.SGD. We've used these flags often earlier in the chapter, but let me give an example of how it works, just to remind you:
>>> import mnist_loader
>>> training_data, validation_data, test_data = \
... mnist_loader.load_data_wrapper()
>>> import network2
>>> net = network2.Network([784, 30, 10], cost=network2.CrossEntropyCost)
>>> net.SGD(training_data, 30, 10, 0.5,
... lmbda = 5.0,
... evaluation_data=validation_data,
... monitor_evaluation_accuracy=True,
... monitor_evaluation_cost=True,
... monitor_training_accuracy=True,
... monitor_training_cost=True)
Here, we're setting the evaluation_data to be the validation_data. But we could also have monitored performance on the test_data or any other data set. We also have four flags telling us to monitor the cost and accuracy on both the evaluation_data and the training_data. Those flags are False by default, but they've been turned on here in order to monitor our Network's performance. Furthermore, network2.py'sNetwork.SGD method returns a four-element tuple representing the results of the monitoring. We can use this as follows:
>>> evaluation_cost, evaluation_accuracy,
... training_cost, training_accuracy = net.SGD(training_data, 30, 10, 0.5,
... lmbda = 5.0,
... evaluation_data=validation_data,
... monitor_evaluation_accuracy=True,
... monitor_evaluation_cost=True,
... monitor_training_accuracy=True,
... monitor_training_cost=True)
So, for example, evaluation_cost will be a 30-element list containing the cost on the evaluation data at the end of each epoch. This sort of information is extremely useful in understanding a network's behaviour. It can, for example, be used to draw graphs showing how the network learns over time. Indeed, that's exactly how I constructed all the graphs earlier in the chapter. Note, however, that if any of the monitoring flags are not set, then the corresponding element in the tuple will be the empty list.
Other additions to the code include a Network.save method, to saveNetwork objects to disk, and a function to load them back in again later. Note that the saving and loading is done using JSON, not Python's pickle or cPickle modules, which are the usual way we save and load objects to and from disk in Python. Using JSON requires more code than pickle or cPickle would. To understand why I've used JSON, imagine that at some time in the future we decided to change our Network class to allow neurons other than sigmoid neurons. To implement that change we'd most likely change the attributes defined in the Network.__init__ method. If we've simply pickled the objects that would cause our load function to fail. Using JSON to do the serialization explicitly makes it easy to ensure that old Networks will still load.
There are many other minor changes in the code for network2.py, but they're all simple variations on network.py. The net result is to expand our 74-line program to a far more capable 152 lines.
Problems
- Modify the code above to implement L1 regularization, and use L1 regularization to classify MNIST digits using a 30 30 hidden neuron network. Can you find a regularization parameter that enables you to do better than running unregularized?
- Take a look at the Network.cost_derivative method in network.py. That method was written for the quadratic cost. How would you rewrite the method for the cross-entropy cost? Can you think of a problem that might arise in the cross-entropy version? In network2.py we've eliminated theNetwork.cost_derivative method entirely, instead incorporating its functionality into the CrossEntropyCost.delta method. How does this solve the problem you've just identified?
How to choose a neural network's hyper-parameters?
Up until now I haven't explained how I've been choosing values for hyper-parameters such as the learning rate, η η, the regularization parameter, λ λ, and so on. I've just been supplying values which work pretty well. In practice, when you're using neural nets to attack a problem, it can be difficult to find good hyper-parameters. Imagine, for example, that we've just been introduced to the MNIST problem, and have begun working on it, knowing nothing at all about what hyper-parameters to use. Let's suppose that by good fortune in our first experiments we choose many of the hyper-parameters in the same way as was done earlier this chapter: 30 hidden neurons, a mini-batch size of 10, training for 30 epochs using the cross-entropy. But we choose a learning rate η=10.0 η=10.0 and regularization parameter λ=1000.0 λ=1000.0. Here's what I saw on one such run:
>>> import mnist_loader
>>> training_data, validation_data, test_data = \
... mnist_loader.load_data_wrapper()
>>> import network2
>>> net = network2.Network([784, 30, 10])
>>> net.SGD(training_data, 30, 10, 10.0, lmbda = 1000.0,
... evaluation_data=validation_data, monitor_evaluation_accuracy=True)
Epoch 0 training complete
Accuracy on evaluation data: 1030 / 10000
Epoch 1 training complete
Accuracy on evaluation data: 990 / 10000
Epoch 2 training complete
Accuracy on evaluation data: 1009 / 10000
...
Epoch 27 training complete
Accuracy on evaluation data: 1009 / 10000
Epoch 28 training complete
Accuracy on evaluation data: 983 / 10000
Epoch 29 training complete
Accuracy on evaluation data: 967 / 10000
Our classification accuracies are no better than chance! Our network is acting as a random noise generator!
"Well, that's easy to fix," you might say, "just decrease the learning rate and regularization hyper-parameters". Unfortunately, you don't a priori know those are the hyper-parameters you need to adjust. Maybe the real problem is that our 30 hidden neuron network will never work well, no matter how the other hyper-parameters are chosen? Maybe we really need at least 100 hidden neurons? Or 300 hidden neurons? Or multiple hidden layers? Or a different approach to encoding the output? Maybe our network is learning, but we need to train for more epochs? Maybe the mini-batches are too small? Maybe we'd do better switching back to the quadratic cost function? Maybe we need to try a different approach to weight initialization? And so on, on and on and on. It's easy to feel lost in hyper-parameter space. This can be particularly frustrating if your network is very large, or uses a lot of training data, since you may train for hours or days or weeks, only to get no result. If the situation persists, it damages your confidence. Maybe neural networks are the wrong approach to your problem? Maybe you should quit your job and take up beekeeping?
In this section I explain some heuristics which can be used to set the hyper-parameters in a neural network. The goal is to help you develop a workflow that enables you to do a pretty good job setting hyper-parameters. Of course, I won't cover everything about hyper-parameter optimization. That's a huge subject, and it's not, in any case, a problem that is ever completely solved, nor is there universal agreement amongst practitioners on the right strategies to use. There's always one more trick you can try to eke out a bit more performance from your network. But the heuristics in this section should get you started.
Broad strategy: When using neural networks to attack a new problem the first challenge is to get any non-trivial learning, i.e., for the network to achieve results better than chance. This can be surprisingly difficult, especially when confronting a new class of problem. Let's look at some strategies you can use if you're having this kind of trouble.
Suppose, for example, that you're attacking MNIST for the first time. You start out enthusiastic, but are a little discouraged when your first network fails completely, as in the example above. The way to go is to strip the problem down. Get rid of all the training and validation images except images which are 0s or 1s. Then try to train a network to distinguish 0s from 1s. Not only is that an inherently easier problem than distinguishing all ten digits, it also reduces the amount of training data by 80 percent, speeding up training by a factor of 5. That enables much more rapid experimentation, and so gives you more rapid insight into how to build a good network.
You can further speed up experimentation by stripping your network down to the simplest network likely to do meaningful learning. If you believe a [784, 10] network can likely do better-than-chance classification of MNIST digits, then begin your experimentation with such a network. It'll be much faster than training a [784, 30, 10] network, and you can build back up to the latter.
You can get another speed up in experimentation by increasing the frequency of monitoring. In network2.py we monitor performance at the end of each training epoch. With 50,000 images per epoch, that means waiting a little while - about ten seconds per epoch, on my laptop, when training a [784, 30, 10] network - before getting feedback on how well the network is learning. Of course, ten seconds isn't very long, but if you want to trial dozens of hyper-parameter choices it's annoying, and if you want to trial hundreds or thousands of choices it starts to get debilitating. We can get feedback more quickly by monitoring the validation accuracy more often, say, after every 1,000 training images. Furthermore, instead of using the full 10,000 image validation set to monitor performance, we can get a much faster estimate using just 100 validation images. All that matters is that the network sees enough images to do real learning, and to get a pretty good rough estimate of performance. Of course, our program network2.py doesn't currently do this kind of monitoring. But as a kludge to achieve a similar effect for the purposes of illustration, we'll strip down our training data to just the first 1,000 MNIST training images. Let's try it and see what happens. (To keep the code below simple I haven't implemented the idea of using only 0 and 1 images. Of course, that can be done with just a little more work.)
>>> net = network2.Network([784, 10])
>>> net.SGD(training_data[:1000], 30, 10, 10.0, lmbda = 1000.0, \
... evaluation_data=validation_data[:100], \
... monitor_evaluation_accuracy=True)
Epoch 0 training complete
Accuracy on evaluation data: 10 / 100
Epoch 1 training complete
Accuracy on evaluation data: 10 / 100
Epoch 2 training complete
Accuracy on evaluation data: 10 / 100
...
We're still getting pure noise! But there's a big win: we're now getting feedback in a fraction of a second, rather than once every ten seconds or so. That means you can more quickly experiment with other choices of hyper-parameter, or even conduct experiments trialling many different choices of hyper-parameter nearly simultaneously.
In the above example I left λ λ as λ=1000.0 λ=1000.0, as we used earlier. But since we changed the number of training examples we should really change λ λ to keep the weight decay the same. That means changing λ λto 20.0 20.0. If we do that then this is what happens:
>>> net = network2.Network([784, 10])
>>> net.SGD(training_data[:1000], 30, 10, 10.0, lmbda = 20.0, \
... evaluation_data=validation_data[:100], \
... monitor_evaluation_accuracy=True)
Epoch 0 training complete
Accuracy on evaluation data: 12 / 100
Epoch 1 training complete
Accuracy on evaluation data: 14 / 100
Epoch 2 training complete
Accuracy on evaluation data: 25 / 100
Epoch 3 training complete
Accuracy on evaluation data: 18 / 100
...
Ahah! We have a signal. Not a terribly good signal, but a signal nonetheless. That's something we can build on, modifying the hyper-parameters to try to get further improvement. Maybe we guess that our learning rate needs to be higher. (As you perhaps realize, that's a silly guess, for reasons we'll discuss shortly, but please bear with me.) So to test our guess we try dialing η η up to 100.0 100.0:
>>> net = network2.Network([784, 10])
>>> net.SGD(training_data[:1000], 30, 10, 100.0, lmbda = 20.0, \
... evaluation_data=validation_data[:100], \
... monitor_evaluation_accuracy=True)
Epoch 0 training complete
Accuracy on evaluation data: 10 / 100
Epoch 1 training complete
Accuracy on evaluation data: 10 / 100
Epoch 2 training complete
Accuracy on evaluation data: 10 / 100
Epoch 3 training complete
Accuracy on evaluation data: 10 / 100
...
That's no good! It suggests that our guess was wrong, and the problem wasn't that the learning rate was too low. So instead we try dialing η η down to η=1.0 η=1.0:
>>> net = network2.Network([784, 10])
>>> net.SGD(training_data[:1000], 30, 10, 1.0, lmbda = 20.0, \
... evaluation_data=validation_data[:100], \
... monitor_evaluation_accuracy=True)
Epoch 0 training complete
Accuracy on evaluation data: 62 / 100
Epoch 1 training complete
Accuracy on evaluation data: 42 / 100
Epoch 2 training complete
Accuracy on evaluation data: 43 / 100
Epoch 3 training complete
Accuracy on evaluation data: 61 / 100
...
That's better! And so we can continue, individually adjusting each hyper-parameter, gradually improving performance. Once we've explored to find an improved value for η η, then we move on to find a good value for λ λ. Then experiment with a more complex architecture, say a network with 10 hidden neurons. Then adjust the values for η η and λ λ again. Then increase to 20 hidden neurons. And then adjust other hyper-parameters some more. And so on, at each stage evaluating performance using our held-out validation data, and using those evaluations to find better and better hyper-parameters. As we do so, it typically takes longer to witness the impact due to modifications of the hyper-parameters, and so we can gradually decrease the frequency of monitoring.
This all looks very promising as a broad strategy. However, I want to return to that initial stage of finding hyper-parameters that enable a network to learn anything at all. In fact, even the above discussion conveys too positive an outlook. It can be immensely frustrating to work with a network that's learning nothing. You can tweak hyper-parameters for days, and still get no meaningful response. And so I'd like to re-emphasize that during the early stages you should make sure you can get quick feedback from experiments. Intuitively, it may seem as though simplifying the problem and the architecture will merely slow you down. In fact, it speeds things up, since you much more quickly find a network with a meaningful signal. Once you've got such a signal, you can often get rapid improvements by tweaking the hyper-parameters. As with many things in life, getting started can be the hardest thing to do.
Okay, that's the broad strategy. Let's now look at some specific recommendations for setting hyper-parameters. I will focus on the learning rate, η η, the L2 regularization parameter, λ λ, and the mini-batch size. However, many of the remarks apply also to other hyper-parameters, including those associated to network architecture, other forms of regularization, and some hyper-parameters we'll meet later in the book, such as the momentum co-efficient.
Learning rate: Suppose we run three MNIST networks with three different learning rates, η=0.025 η=0.025, η=0.25 η=0.25 and η=2.5 η=2.5, respectively. We'll set the other hyper-parameters as for the experiments in earlier sections, running over 30 epochs, with a mini-batch size of 10, and with λ=5.0 λ=5.0. We'll also return to using the full 50,000 50,000 training images. Here's a graph showing the behaviour of the training cost as we train**The graph was generated by multiple_eta.py.:
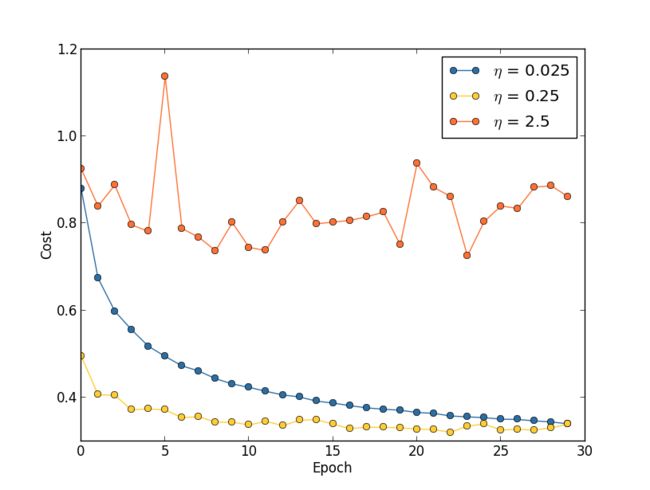
With η=0.025 η=0.025 the cost decreases smoothly until the final epoch. With η=0.25 η=0.25 the cost initially decreases, but after about 20 20 epochs it is near saturation, and thereafter most of the changes are merely small and apparently random oscillations. Finally, with η=2.5 η=2.5 the cost makes large oscillations right from the start. To understand the reason for the oscillations, recall that stochastic gradient descent is supposed to step us gradually down into a valley of the cost function,

However, if η η is too large then the steps will be so large that they may actually overshoot the minimum, causing the algorithm to climb up out of the valley instead. That's likely**This picture is helpful, but it's intended as an intuition-building illustration of what may go on, not as a complete, exhaustive explanation. Briefly, a more complete explanation is as follows: gradient descent uses a first-order approximation to the cost function as a guide to how to decrease the cost. For large η η, higher-order terms in the cost function become more important, and may dominate the behaviour, causing gradient descent to break down. This is especially likely as we approach minima and quasi-minima of the cost function, since near such points the gradient becomes small, making it easier for higher-order terms to dominate behaviour. what's causing the cost to oscillate when η=2.5 η=2.5. When we choose η=0.25 η=0.25 the initial steps do take us toward a minimum of the cost function, and it's only once we get near that minimum that we start to suffer from the overshooting problem. And when we choose η=0.025 η=0.025 we don't suffer from this problem at all during the first 30 30 epochs. Of course, choosing η η so small creates another problem, namely, that it slows down stochastic gradient descent. An even better approach would be to start with η=0.25 η=0.25, train for 20 20 epochs, and then switch to η=0.025 η=0.025. We'll discuss such variable learning rate schedules later. For now, though, let's stick to figuring out how to find a single good value for the learning rate, η η.
With this picture in mind, we can set η η as follows. First, we estimate the threshold value for η η at which the cost on the training data immediately begins decreasing, instead of oscillating or increasing. This estimate doesn't need to be too accurate. You can estimate the order of magnitude by starting with η=0.01 η=0.01. If the cost decreases during the first few epochs, then you should successively try η=0.1,1.0,… η=0.1,1.0,… until you find a value for η η where the cost oscillates or increases during the first few epochs. Alternately, if the cost oscillates or increases during the first few epochs when η=0.01 η=0.01, then try η=0.001,0.0001,… η=0.001,0.0001,… until you find a value for η η where the cost decreases during the first few epochs. Following this procedure will give us an order of magnitude estimate for the threshold value of η η. You may optionally refine your estimate, to pick out the largest value of η η at which the cost decreases during the first few epochs, say η=0.5 η=0.5 or η=0.2 η=0.2 (there's no need for this to be super-accurate). This gives us an estimate for the threshold value of η η.
Obviously, the actual value of η η that you use should be no larger than the threshold value. In fact, if the value of η η is to remain usable over many epochs then you likely want to use a value for η η that is smaller, say, a factor of two below the threshold. Such a choice will typically allow you to train for many epochs, without causing too much of a slowdown in learning.
In the case of the MNIST data, following this strategy leads to an estimate of 0.1 0.1 for the order of magnitude of the threshold value of η η. After some more refinement, we obtain a threshold value η=0.5 η=0.5. Following the prescription above, this suggests using η=0.25 η=0.25 as our value for the learning rate. In fact, I found that using η=0.5 η=0.5worked well enough over 30 30 epochs that for the most part I didn't worry about using a lower value of η η.
This all seems quite straightforward. However, using the training cost to pick η η appears to contradict what I said earlier in this section, namely, that we'd pick hyper-parameters by evaluating performance using our held-out validation data. In fact, we'll use validation accuracy to pick the regularization hyper-parameter, the mini-batch size, and network parameters such as the number of layers and hidden neurons, and so on. Why do things differently for the learning rate? Frankly, this choice is my personal aesthetic preference, and is perhaps somewhat idiosyncratic. The reasoning is that the other hyper-parameters are intended to improve the final classification accuracy on the test set, and so it makes sense to select them on the basis of validation accuracy. However, the learning rate is only incidentally meant to impact the final classification accuracy. Its primary purpose is really to control the step size in gradient descent, and monitoring the training cost is the best way to detect if the step size is too big. With that said, this is a personal aesthetic preference. Early on during learning the training cost usually only decreases if the validation accuracy improves, and so in practice it's unlikely to make much difference which criterion you use.
Use early stopping to determine the number of training epochs: As we discussed earlier in the chapter, early stopping means that at the end of each epoch we should compute the classification accuracy on the validation data. When that stops improving, terminate. This makes setting the number of epochs very simple. In particular, it means that we don't need to worry about explicitly figuring out how the number of epochs depends on the other hyper-parameters. Instead, that's taken care of automatically. Furthermore, early stopping also automatically prevents us from overfitting. This is, of course, a good thing, although in the early stages of experimentation it can be helpful to turn off early stopping, so you can see any signs of overfitting, and use it to inform your approach to regularization.
To implement early stopping we need to say more precisely what it means that the classification accuracy has stopped improving. As we've seen, the accuracy can jump around quite a bit, even when the overall trend is to improve. If we stop the first time the accuracy decreases then we'll almost certainly stop when there are more improvements to be had. A better rule is to terminate if the best classification accuracy doesn't improve for quite some time. Suppose, for example, that we're doing MNIST. Then we might elect to terminate if the classification accuracy hasn't improved during the last ten epochs. This ensures that we don't stop too soon, in response to bad luck in training, but also that we're not waiting around forever for an improvement that never comes.
This no-improvement-in-ten rule is good for initial exploration of MNIST. However, networks can sometimes plateau near a particular classification accuracy for quite some time, only to then begin improving again. If you're trying to get really good performance, the no-improvement-in-ten rule may be too aggressive about stopping. In that case, I suggest using the no-improvement-in-ten rule for initial experimentation, and gradually adopting more lenient rules, as you better understand the way your network trains: no-improvement-in-twenty, no-improvement-in-fifty, and so on. Of course, this introduces a new hyper-parameter to optimize! In practice, however, it's usually easy to set this hyper-parameter to get pretty good results. Similarly, for problems other than MNIST, the no-improvement-in-ten rule may be much too aggressive or not nearly aggressive enough, depending on the details of the problem. However, with a little experimentation it's usually easy to find a pretty good strategy for early stopping.
We haven't used early stopping in our MNIST experiments to date. The reason is that we've been doing a lot of comparisons between different approaches to learning. For such comparisons it's helpful to use the same number of epochs in each case. However, it's well worth modifying network2.py to implement early stopping:
Problem
- Modify network2.py so that it implements early stopping using a no-improvement-in- n n epochs strategy, where n n is a parameter that can be set.
- Can you think of a rule for early stopping other than no-improvement-in- n n? Ideally, the rule should compromise between getting high validation accuracies and not training too long. Add your rule to network2.py, and run three experiments comparing the validation accuracies and number of epochs of training to no-improvement-in- 10 10.
Learning rate schedule: We've been holding the learning rate η ηconstant. However, it's often advantageous to vary the learning rate. Early on during the learning process it's likely that the weights are badly wrong. And so it's best to use a large learning rate that causes the weights to change quickly. Later, we can reduce the learning rate as we make more fine-tuned adjustments to our weights.
How should we set our learning rate schedule? Many approaches are possible. One natural approach is to use the same basic idea as early stopping. The idea is to hold the learning rate constant until the validation accuracy starts to get worse. Then decrease the learning rate by some amount, say a factor of two or ten. We repeat this many times, until, say, the learning rate is a factor of 1,024 (or 1,000) times lower than the initial value. Then we terminate.
A variable learning schedule can improve performance, but it also opens up a world of possible choices for the learning schedule. Those choices can be a headache - you can spend forever trying to optimize your learning schedule. For first experiments my suggestion is to use a single, constant value for the learning rate. That'll get you a good first approximation. Later, if you want to obtain the best performance from your network, it's worth experimenting with a learning schedule, along the lines I've described**A readable recent paper which demonstrates the benefits of variable learning rates in attacking MNIST is Deep, Big, Simple Neural Nets Excel on Handwritten Digit Recognition, by Dan Claudiu Cireșan, Ueli Meier, Luca Maria Gambardella, and Jürgen Schmidhuber (2010)..
Exercise
- Modify network2.py so that it implements a learning schedule that: halves the learning rate each time the validation accuracy satisfies the no-improvement-in- 10 10 rule; and terminates when the learning rate has dropped to 1/128 1/128 of its original value.
The regularization parameter, λ λ: I suggest starting initially with no regularization ( λ=0.0 λ=0.0), and determining a value for η η, as above. Using that choice of η η, we can then use the validation data to select a good value for λ λ. Start by trialling λ=1.0 λ=1.0**I don't have a good principled justification for using this as a starting value. If anyone knows of a good principled discussion of where to start with λ λ, I'd appreciate hearing it ([email protected])., and then increase or decrease by factors of 10 10, as needed to improve performance on the validation data. Once you've found a good order of magnitude, you can fine tune your value of λ λ. That done, you should return and re-optimize η η again.
Exercise
- It's tempting to use gradient descent to try to learn good values for hyper-parameters such as λ λ and η η. Can you think of an obstacle to using gradient descent to determine λ λ? Can you think of an obstacle to using gradient descent to determine η η?
How I selected hyper-parameters earlier in this book: If you use the recommendations in this section you'll find that you get values for η η and λ λ which don't always exactly match the values I've used earlier in the book. The reason is that the book has narrative constraints that have sometimes made it impractical to optimize the hyper-parameters. Think of all the comparisons we've made of different approaches to learning, e.g., comparing the quadratic and cross-entropy cost functions, comparing the old and new methods of weight initialization, running with and without regularization, and so on. To make such comparisons meaningful, I've usually tried to keep hyper-parameters constant across the approaches being compared (or to scale them in an appropriate way). Of course, there's no reason for the same hyper-parameters to be optimal for all the different approaches to learning, so the hyper-parameters I've used are something of a compromise.
As an alternative to this compromise, I could have tried to optimize the heck out of the hyper-parameters for every single approach to learning. In principle that'd be a better, fairer approach, since then we'd see the best from every approach to learning. However, we've made dozens of comparisons along these lines, and in practice I found it too computationally expensive. That's why I've adopted the compromise of using pretty good (but not necessarily optimal) choices for the hyper-parameters.
Mini-batch size: How should we set the mini-batch size? To answer this question, let's first suppose that we're doing online learning, i.e., that we're using a mini-batch size of 1 1.
The obvious worry about online learning is that using mini-batches which contain just a single training example will cause significant errors in our estimate of the gradient. In fact, though, the errors turn out to not be such a problem. The reason is that the individual gradient estimates don't need to be super-accurate. All we need is an estimate accurate enough that our cost function tends to keep decreasing. It's as though you are trying to get to the North Magnetic Pole, but have a wonky compass that's 10-20 degrees off each time you look at it. Provided you stop to check the compass frequently, and the compass gets the direction right on average, you'll end up at the North Magnetic Pole just fine.
Based on this argument, it sounds as though we should use online learning. In fact, the situation turns out to be more complicated than that. In a problem in the last chapter I pointed out that it's possible to use matrix techniques to compute the gradient update for all examples in a mini-batch simultaneously, rather than looping over them. Depending on the details of your hardware and linear algebra library this can make it quite a bit faster to compute the gradient estimate for a mini-batch of (for example) size 100 100, rather than computing the mini-batch gradient estimate by looping over the 100 100 training examples separately. It might take (say) only 50 50 times as long, rather than 100 100 times as long.
Now, at first it seems as though this doesn't help us that much. With our mini-batch of size 100 100 the learning rule for the weights looks like:
With these factors in mind, choosing the best mini-batch size is a compromise. Too small, and you don't get to take full advantage of the benefits of good matrix libraries optimized for fast hardware. Too large and you're simply not updating your weights often enough. What you need is to choose a compromise value which maximizes the speed of learning. Fortunately, the choice of mini-batch size at which the speed is maximized is relatively independent of the other hyper-parameters (apart from the overall architecture), so you don't need to have optimized those hyper-parameters in order to find a good mini-batch size. The way to go is therefore to use some acceptable (but not necessarily optimal) values for the other hyper-parameters, and then trial a number of different mini-batch sizes, scaling η η as above. Plot the validation accuracy versustime (as in, real elapsed time, not epoch!), and choose whichever mini-batch size gives you the most rapid improvement in performance. With the mini-batch size chosen you can then proceed to optimize the other hyper-parameters.
Of course, as you've no doubt realized, I haven't done this optimization in our work. Indeed, our implementation doesn't use the faster approach to mini-batch updates at all. I've simply used a mini-batch size of 10 10 without comment or explanation in nearly all examples. Because of this, we could have sped up learning by reducing the mini-batch size. I haven't done this, in part because I wanted to illustrate the use of mini-batches beyond size 1 1, and in part because my preliminary experiments suggested the speedup would be rather modest. In practical implementations, however, we would most certainly implement the faster approach to mini-batch updates, and then make an effort to optimize the mini-batch size, in order to maximize our overall speed.
Automated techniques: I've been describing these heuristics as though you're optimizing your hyper-parameters by hand. Hand-optimization is a good way to build up a feel for how neural networks behave. However, and unsurprisingly, a great deal of work has been done on automating the process. A common technique isgrid search, which systematically searches through a grid in hyper-parameter space. A review of both the achievements and the limitations of grid search (with suggestions for easily-implemented alternatives) may be found in a 2012 paper**Random search for hyper-parameter optimization, by James Bergstra and Yoshua Bengio (2012). by James Bergstra and Yoshua Bengio. Many more sophisticated approaches have also been proposed. I won't review all that work here, but do want to mention a particularly promising 2012 paper which used a Bayesian approach to automatically optimize hyper-parameters**Practical Bayesian optimization of machine learning algorithms, by Jasper Snoek, Hugo Larochelle, and Ryan Adams.. The code from the paper is publicly available, and has been used with some success by other researchers.
Summing up: Following the rules-of-thumb I've described won't give you the absolute best possible results from your neural network. But it will likely give you a good start and a basis for further improvements. In particular, I've discussed the hyper-parameters largely independently. In practice, there are relationships between the hyper-parameters. You may experiment with η η, feel that you've got it just right, then start to optimize for λ λ, only to find that it's messing up your optimization for η η. In practice, it helps to bounce backward and forward, gradually closing in good values. Above all, keep in mind that the heuristics I've described are rules of thumb, not rules cast in stone. You should be on the lookout for signs that things aren't working, and be willing to experiment. In particular, this means carefully monitoring your network's behaviour, especially the validation accuracy.
The difficulty of choosing hyper-parameters is exacerbated by the fact that the lore about how to choose hyper-parameters is widely spread, across many research papers and software programs, and often is only available inside the heads of individual practitioners. There are many, many papers setting out (sometimes contradictory) recommendations for how to proceed. However, there are a few particularly useful papers that synthesize and distill out much of this lore. Yoshua Bengio has a 2012 paper**Practical recommendations for gradient-based training of deep architectures, by Yoshua Bengio (2012). that gives some practical recommendations for using backpropagation and gradient descent to train neural networks, including deep neural nets. Bengio discusses many issues in much more detail than I have, including how to do more systematic hyper-parameter searches. Another good paper is a 1998 paper**Efficient BackProp, by Yann LeCun, Léon Bottou, Genevieve Orr and Klaus-Robert Müller (1998) by Yann LeCun, Léon Bottou, Genevieve Orr and Klaus-Robert Müller. Both these papers appear in an extremely useful 2012 book that collects many tricks commonly used in neural nets**Neural Networks: Tricks of the Trade, edited by Grégoire Montavon, Geneviève Orr, and Klaus-Robert Müller.. The book is expensive, but many of the articles have been placed online by their respective authors with, one presumes, the blessing of the publisher, and may be located using a search engine.
One thing that becomes clear as you read these articles and, especially, as you engage in your own experiments, is that hyper-parameter optimization is not a problem that is ever completely solved. There's always another trick you can try to improve performance. There is a saying common among writers that books are never finished, only abandoned. The same is also true of neural network optimization: the space of hyper-parameters is so large that one never really finishes optimizing, one only abandons the network to posterity. So your goal should be to develop a workflow that enables you to quickly do a pretty good job on the optimization, while leaving you the flexibility to try more detailed optimizations, if that's important.
The challenge of setting hyper-parameters has led some people to complain that neural networks require a lot of work when compared with other machine learning techniques. I've heard many variations on the following complaint: "Yes, a well-tuned neural network may get the best performance on the problem. On the other hand, I can try a random forest [or SVM or … … insert your own favorite technique] and it just works. I don't have time to figure out just the right neural network." Of course, from a practical point of view it's good to have easy-to-apply techniques. This is particularly true when you're just getting started on a problem, and it may not be obvious whether machine learning can help solve the problem at all. On the other hand, if getting optimal performance is important, then you may need to try approaches that require more specialist knowledge. While it would be nice if machine learning were always easy, there is no a priori reason it should be trivially simple.
Other techniques
Each technique developed in this chapter is valuable to know in its own right, but that's not the only reason I've explained them. The larger point is to familiarize you with some of the problems which can occur in neural networks, and with a style of analysis which can help overcome those problems. In a sense, we've been learning how to think about neural nets. Over the remainder of this chapter I briefly sketch a handful of other techniques. These sketches are less in-depth than the earlier discussions, but should convey some feeling for the diversity of techniques available for use in neural networks.
Variations on stochastic gradient descent
Stochastic gradient descent by backpropagation has served us well in attacking the MNIST digit classification problem. However, there are many other approaches to optimizing the cost function, and sometimes those other approaches offer performance superior to mini-batch stochastic gradient descent. In this section I sketch two such approaches, the Hessian and momentum techniques.
Hessian technique: To begin our discussion it helps to put neural networks aside for a bit. Instead, we're just going to consider the abstract problem of minimizing a cost function C C which is a function of many variables, w=w1,w2,… w=w1,w2,…, so C=C(w) C=C(w). By Taylor's theorem, the cost function can be approximated near a point w w by
- Choose a starting point, w w.
- Update w w to a new point w′=w−H−1∇C w′=w−H−1∇C, where the Hessian H H and ∇C ∇C are computed at w w.
- Update w′ w′ to a new point w′′=w′−H′−1∇′C w′′=w′−H′−1∇′C, where the Hessian H′ H′ and ∇′C ∇′C are computed at w′ w′.
- … …
This approach to minimizing a cost function is known as theHessian technique or Hessian optimization. There are theoretical and empirical results showing that Hessian methods converge on a minimum in fewer steps than standard gradient descent. In particular, by incorporating information about second-order changes in the cost function it's possible for the Hessian approach to avoid many pathologies that can occur in gradient descent. Furthermore, there are versions of the backpropagation algorithm which can be used to compute the Hessian.
If Hessian optimization is so great, why aren't we using it in our neural networks? Unfortunately, while it has many desirable properties, it has one very undesirable property: it's very difficult to apply in practice. Part of the problem is the sheer size of the Hessian matrix. Suppose you have a neural network with 107 107weights and biases. Then the corresponding Hessian matrix will contain 107×107=1014