import numpy as np
import cv2, os
def CVShow(img, title = 'unNamed', max_h = 950, max_w = 1800):
img = np.array(img)
cv2.namedWindow(title, cv2.WINDOW_KEEPRATIO)
H,W = img.shape[:2]
if H > max_h:
ratio = max_h / H
H, W = int(ratio * H), int(ratio * W)
if W > max_w :
ratio = max_w / W
H, W = int(ratio * H), int(ratio * W)
cv2.resizeWindow(title, W, H)
cv2.imshow(title, img) # 自动适应图片大小的,不能缩放
key = cv2.waitKey(0)
if key == ord('s'): # wait for key to write or exit
cv2.imwrite(title+'.jpg', img)
cv2.destroyAllWindows()
return key
def Access_e_ophtha_MA(rootpath):
subdirs, imgpaths = os.listdir(rootpath), []
for subdir in subdirs: # 访问子文件夹
subdirpath = os.path.join(rootpath, subdir)
imgnames = os.listdir(subdirpath)
if 'Thumbs.db' in imgnames:imgnames.remove('Thumbs.db')
for name in imgnames: # 访问子文件夹下的图片
imgpaths.append((os.path.join(subdirpath, name), name))
return imgpaths
def enhance(img):
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8, 8))
return clahe.apply(img)
def Morph_Operate(img):
r1 = cv2.morphologyEx(img, cv2.MORPH_OPEN, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (5, 5)), iterations=1)
R1 = cv2.morphologyEx(r1, cv2.MORPH_CLOSE, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (5, 5)), iterations=1)
r2 = cv2.morphologyEx(R1, cv2.MORPH_OPEN, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (11, 11)), iterations=1)
R2 = cv2.morphologyEx(r2, cv2.MORPH_CLOSE, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (11, 11)), iterations=1)
r3 = cv2.morphologyEx(R2, cv2.MORPH_OPEN, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (23, 23)), iterations=1)
R3 = cv2.morphologyEx(r3, cv2.MORPH_CLOSE, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (23, 23)), iterations=1)
return R3
def Vessels_Mask(f5):
ret, f6 = cv2.threshold(f5, 15, 255, cv2.THRESH_BINARY) # 用来计算 mask
# f7 = cv2.morphologyEx(f6, cv2.MORPH_OPEN, cv2.getStructuringElement(cv2.MORPH_ELLIPSE,(3,3)), iterations=1)
mask = np.ones(f5.shape[:2], dtype="uint8") * 255
_, contours, hierarchy = cv2.findContours(f6, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
for cnt in contours:
if cv2.contourArea(cnt) <= 200:
cv2.drawContours(mask, [cnt], -1, 0, -1)
im = cv2.bitwise_and(f5, f5, mask=mask)
ret, fin = cv2.threshold(im, 15, 255, cv2.THRESH_BINARY_INV)
newfin = cv2.erode(fin, cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (3, 3)), iterations=1)
return newfin
def Vessels_Extract(newfin):
fundus_eroded = cv2.bitwise_not(newfin)
xmask = np.ones(fundus_eroded.shape[:2], dtype="uint8") * 255
_, xcontours, xhierarchy = cv2.findContours(fundus_eroded.copy(), cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE)
for cnt in xcontours:
shape = "unidentified"
peri = cv2.arcLength(cnt, True)
approx = cv2.approxPolyDP(cnt, 0.04 * peri, False)
condition = len(approx) > 4 and cv2.contourArea(cnt) <= 3000 and cv2.contourArea(cnt) >= 100
shape = "circle" if condition else "veins"
if shape == "circle": cv2.drawContours(xmask, [cnt], -1, 0, -1)
finimage = cv2.bitwise_and(fundus_eroded, fundus_eroded, mask=xmask)
blood_vessels = cv2.bitwise_not(finimage)
return blood_vessels
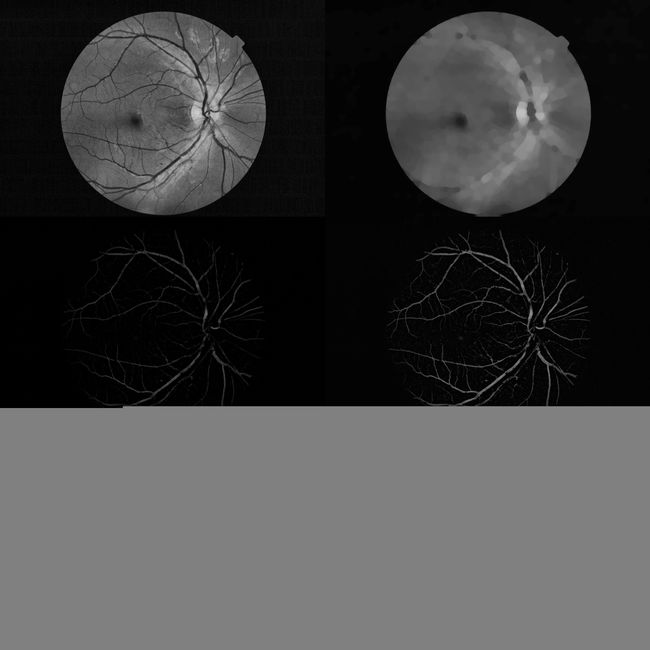
def Vessel_Segmentation_Demo(path):
img = cv2.imread(path)
_, green, _ = cv2.split(img)
contrast_enhanced_green = enhance(green)
morph_contrast_enhanced_green = Morph_Operate(contrast_enhanced_green)
f4 = cv2.subtract(morph_contrast_enhanced_green, contrast_enhanced_green)
f5 = enhance(f4)
newfin = Vessels_Mask(f5)
blood_vessels = Vessels_Extract(newfin)
result1 = np.hstack([contrast_enhanced_green, morph_contrast_enhanced_green])
result2 = np.hstack([f4, f5])
result3 = np.hstack([newfin, blood_vessels])
return np.vstack([result1, result2, result3])
if __name__ == '__main__':
e_ophtha_MA = r'J:\Image Projects\Microaneurysm Detection\Datasets\2013_e_ophtha\e_ophtha_MA'
imgpaths = Access_e_ophtha_MA(e_ophtha_MA + '\MA - CutBlack')
for path,name in imgpaths:
result = Vessel_Segmentation_Demo(path)
key = CVShow(result, name)
if key == ord('b'): break
