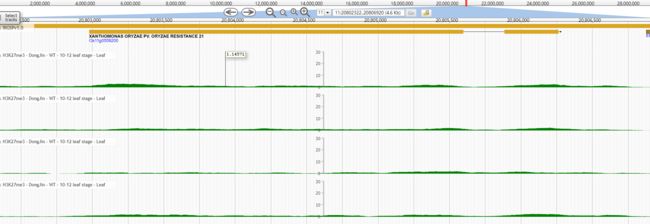
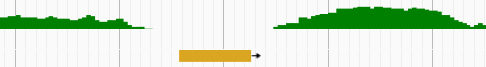
- python—计算学生成绩等级
2111339 彭传月
python
一、打开软件新建窗口输入代码#计算学生成绩等级is_continue='y'whileis_continue=='Y'oris_continue=='y':score=eval(input('请输入学生的成绩:'))ifscore>=90:print('A')elifscore>=80:print('B')elifscore>=70:print('C')elifscore>=60:print('D
- 协议层攻防战:群联AI云防护为何比传统方案更精准?
群联云防护小杜
安全问题汇总人工智能tcp/ip网络协议网络安全
一、四层/七层攻击防御的核心挑战协议层攻击类型传统方案缺陷四层SYNFlood、UDP反射依赖硬件清洗,误封合法流量七层HTTPCC、SQL注入规则静态,无法适应新型攻击二、群联的协议层定制防御技术1.四层协议防护:智能动态指纹技术原理:基于AI分析TCP/UDP流量特征,动态生成协议指纹库,识别伪造源IP的畸形包。文档引用:“防护模块灵活,支持定制版防护模块,适用于非网站业务”。配置示例:#动态
- 无再暴露源站!群联AI云防护IP隐匿方案+防绕过实战
群联云防护小杜
安全问题汇总人工智能tcp/ip网络协议网络安全http服务器
一、IP隐藏的核心原理群联AI云防护通过三层架构实现源站IP深度隐藏:流量入口层:用户访问域名解析至高防CNAME节点(如ai-protect.example.com)智能调度层:基于AI模型动态分配清洗节点,实时更新节点IP池回源层:防护节点通过加密隧道与源站通信,源站仅接受来自群联节点的流量二、IP隐藏配置全流程1.DNS配置(域名指向群联CNAME)#域名DNS记录示例@CNAMEai-pr
- 四层协议攻防手册:从SYN Flood到UDP反射的深度防御
群联云防护小杜
安全问题汇总udp网络网络协议服务器爬虫运维web安全
一、四层协议攻击类型与特征攻击类型协议层特征SYNFloodTCP大量半开连接,SYN_RECV状态堆积UDP反射放大UDP小请求包触发大响应(如NTP、DNS响应)TCP分片攻击TCP发送异常分片耗尽重组资源连接耗尽攻击TCP建立大量空闲连接占用端口资源二、TCP层定制防御方案1.SYNCookie防护(内核参数优化)#启用SYNCookieecho1>/proc/sys/net/ipv4/tc
- CPU占用率飙升至100%:是攻击还是正常现象?
群联云防护小杜
安全问题汇总ddos安全waf服务器cpu占用被攻击
在运维和开发的日常工作中,CPU占用率突然飙升至100%往往是一个令人紧张的信号。这可能意味着服务器正在遭受攻击,但也可能是由于某些正常的、但资源密集型的任务或进程造成的。本文将探讨如何识别和应对服务器的异常CPU占用情况,并通过Python脚本示例,提供一种监控和诊断CPU占用率的方法。一、CPU占用率100%:攻击or正常?1.1攻击迹象持续性高占用:如果CPU占用率长时间保持在100%,且没
- 小程序被黑客攻击,如何防御!
群联云防护小杜
安全问题汇总小程序安全web被攻击阿里云waf
在当今数字化时代,小程序作为连接用户与服务的桥梁,其安全性至关重要。随着小程序生态的日益壮大,也吸引了越来越多的不法分子试图通过各种手段进行攻击,如注入攻击、盗取用户数据、恶意篡改等。为了保护用户隐私和业务安全,开发者必须采取有效的防御措施。本文将深入探讨几种常见的小程序攻击方式及其解决方案,并附带示例代码,以确保您的小程序能够稳健运行。1.SQL注入攻击防范问题描述:攻击者通过在输入字段中插入恶
- Angular中使用Slice管道实现数组特定区间迭代
t0_54program
编程问题解决手册angular.jsjavascript前端个人开发
在Angular开发中,我们经常需要处理数组数据,特别是在模板中迭代这些数据时。然而,标准的*ngFor指令虽然强大,但有时并不能直接满足我们对数组某一特定区间的迭代需求。比如,我们希望跳过数组的第一个元素,或者只迭代数组的前10个元素。今天,我们来看看如何利用Angular提供的Slice管道来实现这个需求。Slice管道介绍Slice管道是一个非常实用的工具,它允许我们从数组或字符串中提取一部
- Angular中`trackBy`函数的独特性与性能优化
t0_54program
编程问题解决手册angular.js前端javascript个人开发
在Angular项目中,优化性能是每一个开发者都需要考虑的问题。特别是在处理大数据量或动态变化的列表时,Angular的trackBy函数成为了我们手中的利器。然而,当我们面对多个列表使用相同trackBy函数时,可能会产生一些疑问:如果这些列表中的项有相同的ID,是否会影响Angular的变更检测?本文将详细探讨trackBy函数在这种情境下的表现及其带来的性能优化。trackBy函数简介tra
- 数据结构-----队列
磨十三
数据结构算法linux
顺序队列(Queue)一、队列核心概念1.基本特性先进先出(FIFO):最早入队的元素最先出队操作限制:队尾(Rear):唯一允许插入的位置队头(Front):唯一允许删除的位置2.顺序队列结构typedefintDATATYPE;typedefstructqueue{DATATYPE*ptr;//存储空间基地址inttlen;//队列总容量inthead;//队头索引inttail;//队尾索引
- 【Python】PDFMiner.six:高效处理PDF文档的Python工具
技术无疆
Pythonpythonpdf开发语言python3.11人工智能数据挖掘机器学习
PDF是一种广泛使用的文件格式,特别适用于呈现固定布局的文档。然而,提取PDF文件中的文本和信息并不总是那么简单。幸好有许多Python库可以帮助我们,其中,PDFMiner.six是一个功能强大、专门用于PDF文档解析的库。⭕️宇宙起点什么是PDFMiner.six?主要功能安装PDFMiner.six♨️核心功能和代码示例1.提取PDF文档的纯文本2.从多个页面提取文本3.提取PDF中的表格内
- RestTemplate和RPC区别
酷爱码
经验分享rpc网络协议网络
RestTemplate是Spring框架中用于进行RESTful风格的HTTP请求的模板类,通常用于与外部服务进行通信。它基于HTTP协议,使用GET、POST、PUT、DELETE等HTTP方法来进行通信,传输的数据通常使用JSON或XML格式。它是一种基于资源的通信方式,通过URL来标识资源。RPC(RemoteProcedureCall)是一种远程过程调用的通信机制,用于不同进程或不同主机
- 从边缘到核心:群联云防护如何重新定义安全加速边界?
群联云防护小杜
安全问题汇总安全分布式ddos前端node.jsudp
一、安全能力的全方位碾压1.协议层深度防护四层防御:动态过滤畸形TCP/UDP包(如SYNFlood),传统CDN仅限速率控制。技术示例:基于AI的协议指纹分析,拦截异常连接模式。七层防御:精准识别业务逻辑攻击(如薅羊毛API调用),CDN仅支持基础URL黑名单。文档引用:“支持基于HTTP头部字段的多条件组合精准访问控制”(产品文档)。2.资源调度与成本优势节点复用:群联共享节点池降低单客户成本
- 25道Python练手题(附详细答案),赶紧收藏!_python题库
字节全栈_rJF
python开发语言
importrandomasrdnumber=rd.randint(0,100)foriinrange(10):choice=int(input("请输入你要猜测的数字:"))ifchoice>number:print("你猜大了")elifchoice0and5*x+3*y+z/3==100:count+=1print("="*60)print(f'第{count}种买法,公鸡买了{x}只,母鸡
- python爱心代码高级
youyouxiong
python开发语言
在Python中,我们可以使用各种方法来绘制一个“爱心”形状。以下是一个使用turtle模块绘制爱心的高级示例。这个示例将使用更复杂的数学公式和图形操作来绘制一个更精致的爱心形状。importturtleimportmath#设置初始状态window=turtle.Screen()window.bgcolor("black")#设置背景色为黑色love=turtle.Turtle()love.sp
- python画一个爱心
戴子雯
python绘画python
大家好这是我的地一篇博客,我要写一个关于python的文章我要用python写一个爱心。不说别的,先看效果效果如下:话不多说,上代码,在这之前要下载python下载这事咱们放在最后现在上代码!!!!!!!!!!!!!!importturtleastt.pensize(2)#笔大小2像素t.pencolor("red")#颜色为红色t.left
- 深入浅出:序列化与反序列化的全面解析
进击的小白菜
一些开发常识开发语言开发常识
文章目录1.引言2.什么是序列化?2.1为什么需要序列化?3.什么是反序列化?3.1反序列化的重要性4.序列化与反序列化的实现4.1JSON(JavaScriptObjectNotation)4.2XML(eXtensibleMarkupLanguage)4.3ProtocolBuffers(Protobuf)4.4MessagePack5.安全性考虑6.性能优化7.结论附录:常见问题解答Q1:什
- brew 安装pip_pip brew wget 安装
weixin_32612253
brew安装pip
终端播放器安装教程从简书上看到一篇,终端实现网易云音乐的文章,并给出了一个github链接.心里有些痒痒,想看看是什么样子,于是尝试安装.安装过程中有些坎坷,记录以便以后查阅.程序实现是用Python写的.安装使用方式仅仅给了三行命令.安装$pipinstallnetease-musicbox$brewinstallmpg123使用$musicbox下载了源码后,不知道该如何安装.三行命令也是莫名
- Centos使用docker搭建Graylog日志平台
moxiaoran5753
centosdockergraylog
日志管理系统有很多,比如ELK,Graylog,Loki+Grafana+Promtail适用场景:1.如果需求复杂,服务器资源不受限制,推荐使用ELK(Logstash+Elasticsearch+Kibana)方案;2.如果需求仅是将不同服务器上的日志采集上来集中展示和检索,且需要一个轻量级的框架,那使用PLG(Promtail+Loki+Grafana)最合适不过了。3.Graylog专注于
- 使用 Baseten 部署和运行机器学习模型的指南
shuoac
机器学习人工智能python
随着机器学习模型在各个行业中的广泛应用,如何高效地部署和运行这些模型成为一个关键问题。本文将介绍如何使用Baseten平台来部署和服务机器学习模型。Baseten是LangChain生态系统中的一个重要提供者,它提供了所需的基础设施来高效地运行模型。无论是开源模型如Llama2和Mistral,还是专有或经过微调的模型,Baseten都能在专用GPU上运行。技术背景介绍Baseten提供了一种不同
- Intent实现参数的传递以及Activity详解
qqmuhua123
Activity是用户唯一可以看得到的东西。几乎所有的activity都与用户进行交互,所以Activity主要负责的就是创建显示窗口,你可以在这些窗口里使用setContentView(View)来显示你自己的UI。onCreate(Bundle)这个方法是初始化activity的地方.最重要的是,你经常需要在这里使用setContentView(int)来设置UI布局所使用的layout资源,
- 从5G向6G演进的三维连接
宋罗世家技术屋
智能科学与技术专栏5G
【摘要】三维连接技术作为地面网络(TN)与非地面网络(NTN)的融合组网技术,既能解决TN空天地海覆盖受限与NTN服务场景受限问题,又能促进后5G(B5G)与6G网络基础设施产业链的健康发展。首先简述了三维连接技术的发展历程,然后重点介绍了未来两年将要完成的5GNTN标准需求、部署结构、空中接口、频谱与终端方面的设计考虑,最后给出了对未来B5G/6G三维连接技术展望,提出了需要全球产学研机构共同研
- MotionLayout(二):MotionLayout是什么?MotionLayout调试技巧、KeyFrame关键帧等等
前期后期
androidkotlin学习
一、MotionLayout是什么?●定位:AndroidJetpack中的高级布局容器,继承自ConstraintLayout。●核心功能:通过状态(State)和过渡(Transition)定义复杂的界面动画,支持手势交互、路径动画等。●优势:简化动画开发流程,替代传统Animator或TransitionManager,适合处理多视图联动、复杂转场效果。1.1应用场景使用MotionLayo
- Android 使用设计模式:装饰者设计模式,对功能进行封装升级,学会可以让我们的代码更加的简洁。
前期后期
设计模式android设计模式
一、前言我遇到什么问题要使用装饰者设计模式?看源码的时候:我们发现明明ui有一个功能,但是在这个ui类找不到,后来发现,这个ui被当做一个参数传递到了一个类里面,后来才在这个类里找到了这个功能。突然醍醐灌顶,这不就是装饰者设计模式吗?写代码的时候:如果我们想给一个功能增加新的东西,可以借助装饰者设计模式来装饰,如果不需要则可以把这个方法去掉,非常的简洁和优雅,并且新增的功能放到了另外一个类里面,也
- Android :实现登录功能的思路
前期后期
android
android的登录功能和前端一样,需要保存登录的用户信息。创建一个工具类//用户工具类,用于管理用户登录状态和用户信息objectAppUserUtil{//常量定义privateconstvalLOGGED_FLAG="logged_flag"//登录状态的键名privateconstvalUSER_INFO="user_info"//用户信息的键名privateconstvalTAG="Ap
- 多级缓存设计实践
MClink
架构缓存
缓存是什么?缓存技术是一种用于加速数据访问的优化策略。它通过将频繁访问的数据存储在高速存储介质(如内存)中,减少对慢速存储设备(如硬盘或远程服务器)的访问次数,从而提升系统的响应速度和性能。缓存的基本原理是:当某个数据被请求时,系统首先检查缓存中是否已存储该数据。如果缓存中存在,则直接返回缓存中的数据,称为“缓存命中”;如果缓存中没有该数据,则从源数据存储(如数据库或远程服务器)中获取数据,并将其
- 探索Google AI聊天模型的集成和使用
qahaj
人工智能python
随着人工智能的飞速发展,GoogleAI的聊天模型提供了强大的自然语言处理能力,可以应用于多种场景中。本文将为你介绍如何通过GoogleAI和LangChain库来使用这些聊天模型。技术背景介绍GoogleAI提供了一系列强大的聊天模型,这些模型具备不同的功能和参数设置。它们不仅可以通过GoogleAI服务访问,还可以通过GoogleCloudVertexAI以企业级功能使用。在本文中,我们将重点
- Windows 图形显示驱动开发-WDDM 2.7功能- 支持跨适配器资源扫描 (CASO)
程序员王马
windows图形显示驱动开发windows驱动开发
Microsoft计算驱动程序模型概述在Windows10版本1903(WDDM2.6)及更高版本中,Microsoft计算驱动程序模型(MCDM)可用于为支持仅计算功能的设备编写驱动程序。MCDM驱动程序或仅计算驱动程序是Windows显示驱动程序模型2.0+(WDDM)的缩减子集。在WDDM术语中,驱动程序必须将自身播发为“仅呈现”设备,而无需显示功能。“呈现设备”的内核支持很灵活,因为设备执
- 一文读懂 Linux 下 Docker 搭建及简单应用
Waitccy
linuxdocker运维服务器
一、引言在Linux系统的运维与开发场景中,Docker凭借其高效的容器化技术,极大地简化了应用部署与管理流程。它打破了传统环境配置的复杂性,实现应用及其依赖的封装,确保在不同环境中稳定运行。本文将详细介绍在Linux系统下搭建Docker的步骤,并通过几个简单应用示例,带你快速上手Docker。二、Linux下Docker搭建(一)准备工作系统要求:建议使用主流的Linux发行版,如Ubuntu
- 扫地机高增长神话破灭!科沃斯、石头科技艰难 “破冰”!
liukuang110
科技
扫地机器人赛道太冷,陆续有企业倒在寒风里。先是,老牌研发商广东宝乐机器人宣布破产重整;曾获得腾讯和红杉资本大额融资,并邀请罗永浩代言的“追光”品牌,也在短短两年内宣告失败。就连雷军投资、小米生态链孵化的睿米科技,也发布了停止运营的通告。头部玩家近况亦不乐观。以科技创新而闻名的科沃斯业绩大幅下滑,在过去几个月中股价的剧烈下跌,引发了市场的高度关注与深刻反思。另一头部玩家石头科技,毛利率下滑、存货周转
- 线程中run方法与start方法的差别
夜君客
java开发语言
run()方法run()方法是Runnable接口中定义的方法,Thread类实现了Runnable接口。当你直接调用run()方法时,它会在当前线程中执行,而不会启动一个新的线程。也就是说,run()方法只是一个普通的方法调用,不会产生多线程的效果。start()方法start()方法用于启动一个新的线程。当你调用start()方法时,JVM会创建一个新的线程,并在这个新线程中调用run()方法
- redis学习笔记——不仅仅是存取数据
Everyday都不同
returnSourceexpire/delincr/lpush数据库分区redis
最近项目中用到比较多redis,感觉之前对它一直局限于get/set数据的层面。其实作为一个强大的NoSql数据库产品,如果好好利用它,会带来很多意想不到的效果。(因为我搞java,所以就从jedis的角度来补充一点东西吧。PS:不一定全,只是个人理解,不喜勿喷)
1、关于JedisPool.returnSource(Jedis jeids)
这个方法是从red
- SQL性能优化-持续更新中。。。。。。
atongyeye
oraclesql
1 通过ROWID访问表--索引
你可以采用基于ROWID的访问方式情况,提高访问表的效率, , ROWID包含了表中记录的物理位置信息..ORACLE采用索引(INDEX)实现了数据和存放数据的物理位置(ROWID)之间的联系. 通常索引提供了快速访问ROWID的方法,因此那些基于索引列的查询就可以得到性能上的提高.
2 共享SQL语句--相同的sql放入缓存
3 选择最有效率的表
- [JAVA语言]JAVA虚拟机对底层硬件的操控还不完善
comsci
JAVA虚拟机
如果我们用汇编语言编写一个直接读写CPU寄存器的代码段,然后利用这个代码段去控制被操作系统屏蔽的硬件资源,这对于JVM虚拟机显然是不合法的,对操作系统来讲,这样也是不合法的,但是如果是一个工程项目的确需要这样做,合同已经签了,我们又不能够这样做,怎么办呢? 那么一个精通汇编语言的那种X客,是否在这个时候就会发生某种至关重要的作用呢?
&n
- lvs- real
男人50
LVS
#!/bin/bash
#
# Script to start LVS DR real server.
# description: LVS DR real server
#
#. /etc/rc.d/init.d/functions
VIP=10.10.6.252
host='/bin/hostname'
case "$1" in
sta
- 生成公钥和私钥
oloz
DSA安全加密
package com.msserver.core.util;
import java.security.KeyPair;
import java.security.PrivateKey;
import java.security.PublicKey;
import java.security.SecureRandom;
public class SecurityUtil {
- UIView 中加入的cocos2d,背景透明
374016526
cocos2dglClearColor
要点是首先pixelFormat:kEAGLColorFormatRGBA8,必须有alpha层才能透明。然后view设置为透明glView.opaque = NO;[director setOpenGLView:glView];[self.viewController.view setBackgroundColor:[UIColor clearColor]];[self.viewControll
- mysql常用命令
香水浓
mysql
连接数据库
mysql -u troy -ptroy
备份表
mysqldump -u troy -ptroy mm_database mm_user_tbl > user.sql
恢复表(与恢复数据库命令相同)
mysql -u troy -ptroy mm_database < user.sql
备份数据库
mysqldump -u troy -ptroy
- 我的架构经验系列文章 - 后端架构 - 系统层面
agevs
JavaScriptjquerycsshtml5
系统层面:
高可用性
所谓高可用性也就是通过避免单独故障加上快速故障转移实现一旦某台物理服务器出现故障能实现故障快速恢复。一般来说,可以采用两种方式,如果可以做业务可以做负载均衡则通过负载均衡实现集群,然后针对每一台服务器进行监控,一旦发生故障则从集群中移除;如果业务只能有单点入口那么可以通过实现Standby机加上虚拟IP机制,实现Active机在出现故障之后虚拟IP转移到Standby的快速
- 利用ant进行远程tomcat部署
aijuans
tomcat
在javaEE项目中,需要将工程部署到远程服务器上,如果部署的频率比较高,手动部署的方式就比较麻烦,可以利用Ant工具实现快捷的部署。这篇博文详细介绍了ant配置的步骤(http://www.cnblogs.com/GloriousOnion/archive/2012/12/18/2822817.html),但是在tomcat7以上不适用,需要修改配置,具体如下:
1.配置tomcat的用户角色
- 获取复利总收入
baalwolf
获取
public static void main(String args[]){
int money=200;
int year=1;
double rate=0.1;
&
- eclipse.ini解释
BigBird2012
eclipse
大多数java开发者使用的都是eclipse,今天感兴趣去eclipse官网搜了一下eclipse.ini的配置,供大家参考,我会把关键的部分给大家用中文解释一下。还是推荐有问题不会直接搜谷歌,看官方文档,这样我们会知道问题的真面目是什么,对问题也有一个全面清晰的认识。
Overview
1、Eclipse.ini的作用
Eclipse startup is controlled by th
- AngularJS实现分页功能
bijian1013
JavaScriptAngularJS分页
对于大多数web应用来说显示项目列表是一种很常见的任务。通常情况下,我们的数据会比较多,无法很好地显示在单个页面中。在这种情况下,我们需要把数据以页的方式来展示,同时带有转到上一页和下一页的功能。既然在整个应用中这是一种很常见的需求,那么把这一功能抽象成一个通用的、可复用的分页(Paginator)服务是很有意义的。
&nbs
- [Maven学习笔记三]Maven archetype
bit1129
ArcheType
archetype的英文意思是原型,Maven archetype表示创建Maven模块的模版,比如创建web项目,创建Spring项目等等.
mvn archetype提供了一种命令行交互式创建Maven项目或者模块的方式,
mvn archetype
1.在LearnMaven-ch03目录下,执行命令mvn archetype:gener
- 【Java命令三】jps
bit1129
Java命令
jps很简单,用于显示当前运行的Java进程,也可以连接到远程服务器去查看
[hadoop@hadoop bin]$ jps -help
usage: jps [-help]
jps [-q] [-mlvV] [<hostid>]
Definitions:
<hostid>: <hostname>[:
- ZABBIX2.2 2.4 等各版本之间的兼容性
ronin47
zabbix更新很快,从2009年到现在已经更新多个版本,为了使用更多zabbix的新特性,随之而来的便是升级版本,zabbix版本兼容性是必须优先考虑的一点 客户端AGENT兼容
zabbix1.x到zabbix2.x的所有agent都兼容zabbix server2.4:如果你升级zabbix server,客户端是可以不做任何改变,除非你想使用agent的一些新特性。 Zabbix代理(p
- unity 3d还是cocos2dx哪个适合游戏?
brotherlamp
unity自学unity教程unity视频unity资料unity
unity 3d还是cocos2dx哪个适合游戏?
问:unity 3d还是cocos2dx哪个适合游戏?
答:首先目前来看unity视频教程因为是3d引擎,目前对2d支持并不完善,unity 3d 目前做2d普遍两种思路,一种是正交相机,3d画面2d视角,另一种是通过一些插件,动态创建mesh来绘制图形单元目前用的较多的是2d toolkit,ex2d,smooth moves,sm2,
- 百度笔试题:一个已经排序好的很大的数组,现在给它划分成m段,每段长度不定,段长最长为k,然后段内打乱顺序,请设计一个算法对其进行重新排序
bylijinnan
java算法面试百度招聘
import java.util.Arrays;
/**
* 最早是在陈利人老师的微博看到这道题:
* #面试题#An array with n elements which is K most sorted,就是每个element的初始位置和它最终的排序后的位置的距离不超过常数K
* 设计一个排序算法。It should be faster than O(n*lgn)。
- 获取checkbox复选框的值
chiangfai
checkbox
<title>CheckBox</title>
<script type = "text/javascript">
doGetVal: function doGetVal()
{
//var fruitName = document.getElementById("apple").value;//根据
- MySQLdb用户指南
chenchao051
mysqldb
原网页被墙,放这里备用。 MySQLdb User's Guide
Contents
Introduction
Installation
_mysql
MySQL C API translation
MySQL C API function mapping
Some _mysql examples
MySQLdb
- HIVE 窗口及分析函数
daizj
hive窗口函数分析函数
窗口函数应用场景:
(1)用于分区排序
(2)动态Group By
(3)Top N
(4)累计计算
(5)层次查询
一、分析函数
用于等级、百分点、n分片等。
函数 说明
RANK() &nbs
- PHP ZipArchive 实现压缩解压Zip文件
dcj3sjt126com
PHPzip
PHP ZipArchive 是PHP自带的扩展类,可以轻松实现ZIP文件的压缩和解压,使用前首先要确保PHP ZIP 扩展已经开启,具体开启方法就不说了,不同的平台开启PHP扩增的方法网上都有,如有疑问欢迎交流。这里整理一下常用的示例供参考。
一、解压缩zip文件 01 02 03 04 05 06 07 08 09 10 11
- 精彩英语贺词
dcj3sjt126com
英语
I'm always here
我会一直在这里支持你
&nb
- 基于Java注解的Spring的IoC功能
e200702084
javaspringbeanIOCOffice
- java模拟post请求
geeksun
java
一般API接收客户端(比如网页、APP或其他应用服务)的请求,但在测试时需要模拟来自外界的请求,经探索,使用HttpComponentshttpClient可模拟Post提交请求。 此处用HttpComponents的httpclient来完成使命。
import org.apache.http.HttpEntity ;
import org.apache.http.HttpRespon
- Swift语法之 ---- ?和!区别
hongtoushizi
?swift!
转载自: http://blog.sina.com.cn/s/blog_71715bf80102ux3v.html
Swift语言使用var定义变量,但和别的语言不同,Swift里不会自动给变量赋初始值,也就是说变量不会有默认值,所以要求使用变量之前必须要对其初始化。如果在使用变量之前不进行初始化就会报错:
var stringValue : String
//
- centos7安装jdk1.7
jisonami
jdkcentos
安装JDK1.7
步骤1、解压tar包在当前目录
[root@localhost usr]#tar -xzvf jdk-7u75-linux-x64.tar.gz
步骤2:配置环境变量
在etc/profile文件下添加
export JAVA_HOME=/usr/java/jdk1.7.0_75
export CLASSPATH=/usr/java/jdk1.7.0_75/lib
- 数据源架构模式之数据映射器
home198979
PHP架构数据映射器datamapper
前面分别介绍了数据源架构模式之表数据入口、数据源架构模式之行和数据入口数据源架构模式之活动记录,相较于这三种数据源架构模式,数据映射器显得更加“高大上”。
一、概念
数据映射器(Data Mapper):在保持对象和数据库(以及映射器本身)彼此独立的情况下,在二者之间移动数据的一个映射器层。概念永远都是抽象的,简单的说,数据映射器就是一个负责将数据映射到对象的类数据。
&nb
- 在Python中使用MYSQL
pda158
mysqlpython
缘由 近期在折腾一个小东西须要抓取网上的页面。然后进行解析。将结果放到
数据库中。 了解到
Python在这方面有优势,便选用之。 由于我有台
server上面安装有
mysql,自然使用之。在进行数据库的这个操作过程中遇到了不少问题,这里
记录一下,大家共勉。
python中mysql的调用
百度之后能够通过MySQLdb进行数据库操作。
- 单例模式
hxl1988_0311
java单例设计模式单件
package com.sosop.designpattern.singleton;
/*
* 单件模式:保证一个类必须只有一个实例,并提供全局的访问点
*
* 所以单例模式必须有私有的构造器,没有私有构造器根本不用谈单件
*
* 必须考虑到并发情况下创建了多个实例对象
* */
/**
* 虽然有锁,但是只在第一次创建对象的时候加锁,并发时不会存在效率
- 27种迹象显示你应该辞掉程序员的工作
vipshichg
工作
1、你仍然在等待老板在2010年答应的要提拔你的暗示。 2、你的上级近10年没有开发过任何代码。 3、老板假装懂你说的这些技术,但实际上他完全不知道你在说什么。 4、你干完的项目6个月后才部署到现场服务器上。 5、时不时的,老板在检查你刚刚完成的工作时,要求按新想法重新开发。 6、而最终这个软件只有12个用户。 7、时间全浪费在办公室政治中,而不是用在开发好的软件上。 8、部署前5分钟才开始测试。