机器学习入门04——糖尿病数据预测
利用Logistic回归技术实现糖尿病发病预测
数据说明
数据说明: Pima Indians Diabetes Data Set(皮马印第安人糖尿病数据集) 根据现有的医疗信息预测5年内皮马印第安人糖尿病发作的概率。
数据链接:https://archive.ics.uci.edu/ml/datasets/Pima+Indians+Diabetes
p.s.: Kaggle也有一个Practice Fusion Diabetes Classification任务,可以试试:)
https://www.kaggle.com/c/pf2012-diabetes
1)文件说明
pima-indians-diabetes.csv:数据文件
2)字段说明
数据集共9个字段:
pregnants:怀孕次数
Plasma_glucose_concentration:口服葡萄糖耐量试验中2小时后的血浆葡萄糖浓度
blood_pressure:舒张压,单位:mm Hg
Triceps_skin_fold_thickness:三头肌皮褶厚度,单位:mm
serum_insulin:餐后血清胰岛素,单位:mm
BMI:体重指数(体重(公斤)/ 身高(米)^2)
Diabetes_pedigree_function:糖尿病家系作用
Age:年龄
Target:标签, 0表示不发病,1表示发病
第一步 特征工程
对于原始数据的处理使用了一下几个方法:
- 使用中值补充缺失数据
- 使用StandardScaler()对数据进行标准化处理
- 保存为csv格式
import numpy as np
import pandas as pd
#input data
train = pd.read_csv("pima-indians-diabetes.csv")
print(train.head())
#查看缺失值较多的数据统计
NaN_col_names = ['Plasma_glucose_concentration','blood_pressure','Triceps_skin_fold_thickness','serum_insulin','BMI']
train[NaN_col_names] = train[NaN_col_names].replace(0, np.NaN)
print(train.isnull().sum())
#中值补充确实值
medians = train.median()
train = train.fillna(medians)
print(train.isnull().sum())
# get labels
y_train = train['Target']
X_train = train.drop(["Target"], axis=1)
#用于保存特征工程之后的结果
feat_names = X_train.columns
# 数据标准化
from sklearn.preprocessing import StandardScaler
# 初始化特征的标准化器
ss_X = StandardScaler()
# 分别对训练和测试数据的特征进行标准化处理
X_train = ss_X.fit_transform(X_train)
#存为csv格式
X_train = pd.DataFrame(columns = feat_names, data = X_train)
train = pd.concat([X_train, y_train], axis = 1)
train.to_csv('FE_pima-indians-diabetes.csv',index = False,header=True)
print(train.head())
第二步 训练模型
训练模型主要分为以下几个步骤:
- 导入数据
- 分别使用L1正则和L2正则训练模型
- 使用5折交叉验证
- 使用log似然损失和正确率对模型进行超参数调优
- 绘制CV误差曲线
# 首先 import 必要的模块
import pandas as pd
import numpy as np
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
from sklearn.model_selection import GridSearchCV
from sklearn.linear_model import LogisticRegression
#%matplotlib inline
# 读取数据
train = pd.read_csv('FE_pima-indians-diabetes.csv')
y_train = train['Target']
X_train = train.drop(["Target"], axis=1)
#保存特征名字以备后用(可视化)
feat_names = X_train.columns
#需要调优的参数
# 请尝试将L1正则和L2正则分开,并配合合适的优化求解算法(slover)
penaltys = ['l1','l2']
#训练数据多,C可以大一点(更多相信数据)
Cs = [0.01, 0.1, 1, 10, 100, 1000, 10000]
tuned_parameters = dict(penalty = penaltys, C = Cs)#组合调优参数
lr_penalty= LogisticRegression()
#grid= GridSearchCV(lr_penalty, tuned_parameters,cv=5, scoring='neg_log_loss')#log似然损失
grid= GridSearchCV(lr_penalty, tuned_parameters,cv=5, scoring='accuracy')#正确率
grid.fit(X_train,y_train)
# examine the best model
print(-grid.best_score_)#打印模型参数
print(grid.best_params_)
#绘制CV误差曲线分析模型
# plot CV误差曲线
test_means = grid.cv_results_[ 'mean_test_score' ]
test_stds = grid.cv_results_[ 'std_test_score' ]
train_means = grid.cv_results_[ 'mean_train_score' ]
train_stds = grid.cv_results_[ 'std_train_score' ]
# plot results
n_Cs = len(Cs)
number_penaltys = len(penaltys)
test_scores = np.array(test_means).reshape(n_Cs,number_penaltys)
train_scores = np.array(train_means).reshape(n_Cs,number_penaltys)
test_stds = np.array(test_stds).reshape(n_Cs,number_penaltys)
train_stds = np.array(train_stds).reshape(n_Cs,number_penaltys)
x_axis = np.log10(Cs)
for i, value in enumerate(penaltys):
#pyplot.plot(log(Cs), test_scores[i], label= 'penalty:' + str(value))
plt.errorbar(x_axis, -test_scores[:,i], yerr=test_stds[:,i] ,label = penaltys[i] +' Test')
#plt.errorbar(x_axis, -train_scores[:,i], yerr=train_stds[:,i] ,label = penaltys[i] +' Train')
plt.legend()
plt.xlabel( 'log(C)' )
plt.ylabel( 'logloss' )
plt.savefig('LogisticGridSearchCV_C.png' )
plt.show()
import cPickle
cPickle.dump(grid.best_estimator_, open("FE_pima-indians-diabetes", 'wb'))
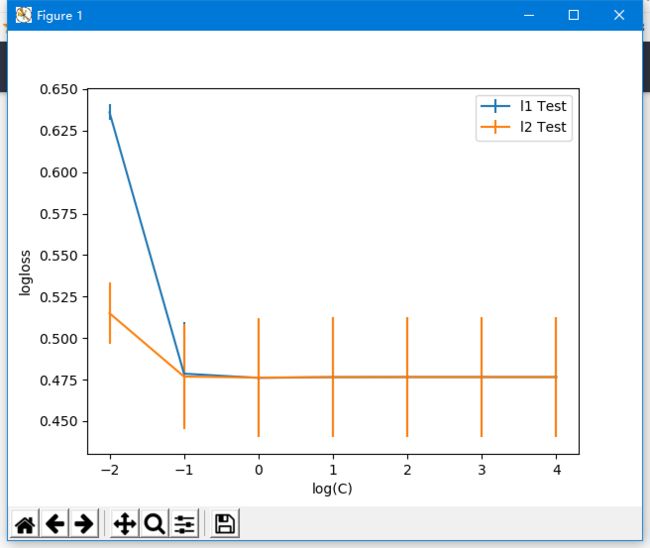
使用log似然损失对Logistic回归模型的正则超参数调优
参数结果:
0.47602775434873434
{'C': 1, 'penalty': 'l1'}
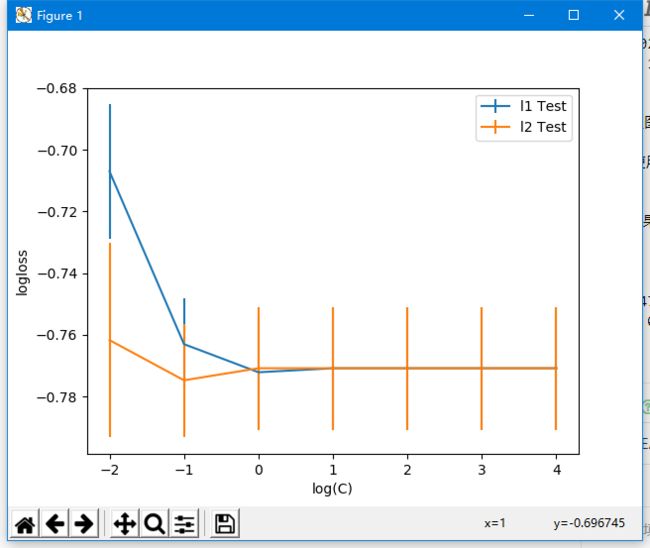
使用log似然损失对Logistic回归模型的正则超参数调优
参数结果:
-0.7747395833333334
{'C': 0.1, 'penalty': 'l2'}
可以看到,使用正确率和l2正则对Logistic回归模型的正则超参数调优可以使损失函数最小
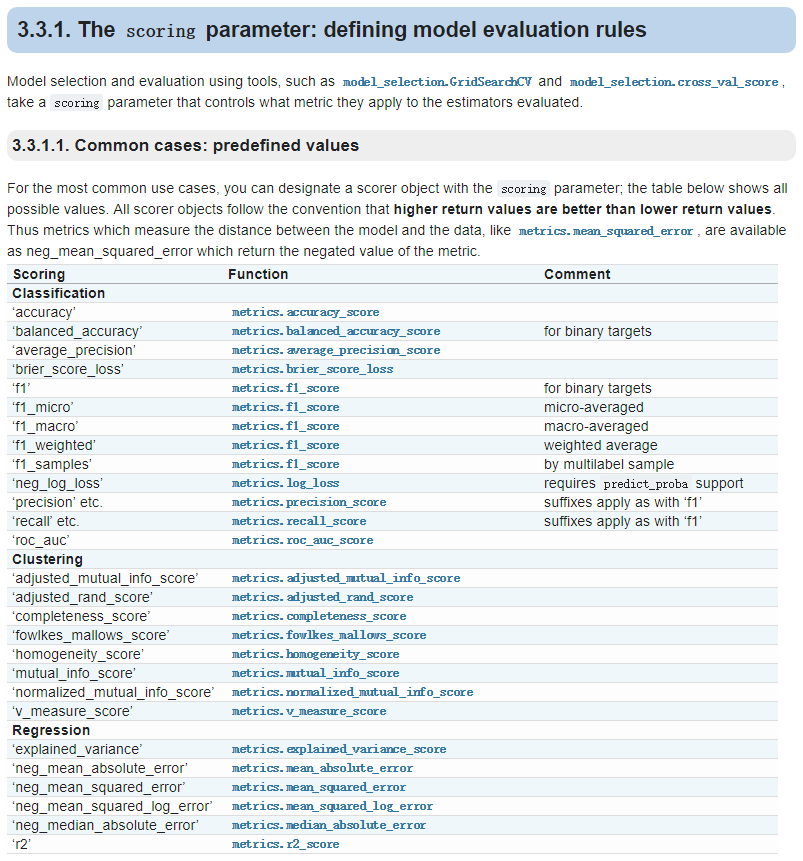
GridSearchCV()中scoring的值可以选用一下参数:
点击这里可以到官网查看!!!
项目源代码:糖尿病发病率预测