用circlize包绘制circos-plot
作者简介Introduction
taoyan:伪码农,R语言爱好者,爱开源。
个人博客: https://ytlogos.github.io/
公众号:生信大讲堂
往期回顾
R语言可视化学习笔记之相关矩阵可视化包ggcorrplot
R语言可视化学习笔记之ggrepel包
R语言学习笔记之相关性矩阵分析及其可视化
ggplot2学习笔记系列之利用ggplot2绘制误差棒及显著性标记
ggplot2学习笔记系列之主题(theme)设置
circlize包
circlize包在德国癌症中心的华人博士Zuguang Gu开发的,有兴趣的可以去看看他的Github主页(https://github.com/jokergoo)。这个包有两个文档,一个是介绍基本原理的绘制简单圈圈图的,也是本次要介绍的。另外一份文档专门介绍基因组数据绘制圈圈图Genomic Circos Plot,我自己还没看完,下次再介绍。
根据我的学习发现这个包与ggplot2很相似,也是先创建一个图层,然后不断的添加图形元素(point、line、bar等),这些简单的图形元素都有circos.这个前缀进行绘制,比如要绘制点,则用circos.points()。具体的下面一一介绍。
用circlize绘制圈圈图
照例,没有安装这个包的先安装:install.packages("circlize")或者devtools::install_github("jokergoo/circlize")。
绘图第一步是先初始化(circos.initialize),接下来绘制track,再添加基本元素。需要提一下的是,由于circlize绘制图是不断叠加的,因此如果我们一大段代码下来我们只能看到最终的图形,这里为了演示每端代码的结果,所以每次我都得初始化以及circlize.clear。
library(circlize)
# 简单创建一个数据集
set.seed(999)
n <- 1000
a <- data.frame(factors = sample(letters[1:8], n, replace = TRUE), x = rnorm(n), y = runif(n))
绘制第一个track
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
#初始化,factors来控制track数目,初始化里只有x, 没有y。这一步相当于ggplot()
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) {
circos.axis()})
col <- rep(c("#FF0000", "#00FF00"), 4) #自定义一下颜色
# 这里先解释一下,一个track有好几个cell,具体数目由factors决定的,向本数据集中factors有八个,因此绘制一个track,其包含八个cell。含有前缀circos.track的函数会在所有的cel里添加基本元素,而只有前缀circos.的函数可以在特定的track、cell里添加基本元素。具体看下演示。
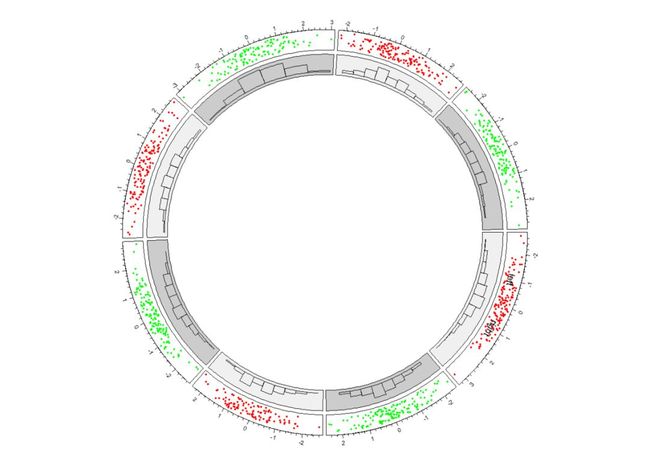
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5) #所有的cell里都绘制点图
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1) #在track 1中的标记为a的cell里添加
textcircos.text(1, 0.5, "right", sector.index = "a")
circos.clear()
接下来绘制第二个track
circos.trackHist添加柱状图,由于柱状图相对高级一点,因此circos.trackHist会自动创建一个track,无需我们circos.trackPlotRegion进行创建。
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) {
circos.axis()})
col <- rep(c("#FF0000", "#00FF00"), 4)
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
bg.col <- rep(c("#EFEFEF", "#CCCCCC"), 4)
circos.trackHist(a$factors, a$x, bg.col = bg.col, col = NA)
circos.clear()
创建第三个track
这里又得提一下,当我们绘制多个track时,我们添加基本元素时要指定添加到哪个track(track.index指定)、哪个cell(sector.index指定)里,如果不指定,那么将默认track是我们刚刚创建的那个。track.index、sector.index等参数可以通过get.cell.meta.data函数获取。
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) {
circos.axis()})
col <- rep(c("#FF0000", "#00FF00"), 4)
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
bg.col <- rep(c("#EFEFEF", "#CCCCCC"), 4)
circos.trackHist(a$factors, a$x, bg.col = bg.col, col = NA)
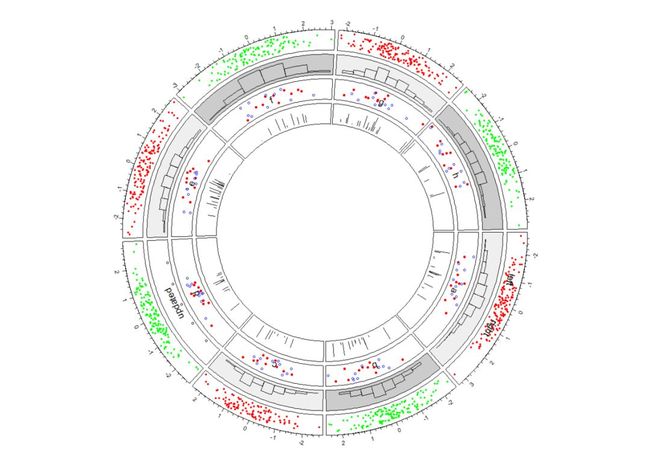
circos.trackPlotRegion(factors = a$factors, x = a$x, y = a$y,
panel.fun = function(x, y) {
grey = c("#FFFFFF", "#CCCCCC", "#999999")
sector.index = get.cell.meta.data("sector.index") #这个是第三个track,因为我们刚刚创建,这里这一步不用也可。
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), sector.index)
circos.points(x[1:10], y[1:10], col = "red", pch = 16, cex = 0.6)
circos.points(x[11:20], y[11:20], col = "blue", cex = 0.6)})
circos.clear()
实际操作中我们常常会更新数据或者想更新图形,这是可以通过circos.updatePlotRegion函数在特定的track、cell里(先删除再添加)update,下面我们将通过circos.updatePlotRegion函数先删除track 2、sector d中的图形元素再添加点图。
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) {
circos.axis()})col <- rep(c("#FF0000", "#00FF00"), 4)
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
bg.col <- rep(c("#EFEFEF", "#CCCCCC"), 4)
circos.trackHist(a$factors, a$x, bg.col = bg.col, col = NA)
circos.trackPlotRegion(factors = a$factors, x = a$x, y = a$y,
panel.fun = function(x, y) {
grey = c("#FFFFFF", "#CCCCCC", "#999999")
sector.index = get.cell.meta.data("sector.index")
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), sector.index)
circos.points(x[1:10], y[1:10], col = "red", pch = 16, cex = 0.6)
circos.points(x[11:20], y[11:20], col = "blue", cex = 0.6)})
# update第2个track中标记为d的sector
circos.updatePlotRegion(sector.index = "d", track.index = 2)
circos.points(x = -2:2, y = rep(0, 5))
xlim <- get.cell.meta.data("xlim")
ylim <- get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), "updated")
circos.clear()
接下来绘制第四个track
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) { circos.axis()})
col <- rep(c("#FF0000", "#00FF00"), 4)
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
bg.col <- rep(c("#EFEFEF", "#CCCCCC"), 4)
circos.trackHist(a$factors, a$x, bg.col = bg.col, col = NA)
circos.trackPlotRegion(factors = a$factors, x = a$x, y = a$y,
panel.fun = function(x, y) {
grey = c("#FFFFFF", "#CCCCCC", "#999999")
sector.index = get.cell.meta.data("sector.index")
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), sector.index)
circos.points(x[1:10], y[1:10], col = "red", pch = 16, cex = 0.6)
circos.points(x[11:20], y[11:20], col = "blue", cex = 0.6)})
# update第2个track中标记为d的sector
circos.updatePlotRegion(sector.index = "d", track.index = 2)
circos.points(x = -2:2, y = rep(0, 5))
xlim <- get.cell.meta.data("xlim")
ylim <- get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), "updated")
circos.clear()
circos.trackPlotRegion(factors = a$factors, y = a$y)
circos.trackLines(a$factors[1:100], a$x[1:100], a$y[1:100], type = "h")
接下来添加links,links可以是point到point、point到interval、interval到interval
par(mar = c(1, 1, 1, 1), lwd = 0.1, cex = 0.6)
circos.par(track.height = 0.1)
circos.initialize(factors = a$factors, x = a$x)
circos.trackPlotRegion(factors = a$factors, y = a$y,
panel.fun = function(x, y) { circos.axis()})
col <- rep(c("#FF0000", "#00FF00"), 4)
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
circos.trackPoints(a$factors, a$x, a$y, col = col, pch = 16, cex = 0.5)
circos.text(-1, 0.5, "left", sector.index = "a", track.index = 1)
circos.text(1, 0.5, "right", sector.index = "a")
bg.col <- rep(c("#EFEFEF", "#CCCCCC"), 4)
circos.trackHist(a$factors, a$x, bg.col = bg.col, col = NA)
circos.trackPlotRegion(factors = a$factors, x = a$x, y = a$y,
panel.fun = function(x, y) {
grey = c("#FFFFFF", "#CCCCCC", "#999999")
sector.index = get.cell.meta.data("sector.index")
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), sector.index)
circos.points(x[1:10], y[1:10], col = "red", pch = 16, cex = 0.6)
circos.points(x[11:20], y[11:20], col = "blue", cex = 0.6)})
# update第2个track中标记为d的sector
circos.updatePlotRegion(sector.index = "d", track.index = 2)
circos.points(x = -2:2, y = rep(0, 5))
xlim <- get.cell.meta.data("xlim")
ylim <- get.cell.meta.data("ylim")
circos.text(mean(xlim), mean(ylim), "updated")
circos.clear()
circos.trackPlotRegion(factors = a$factors, y = a$y)
circos.trackLines(a$factors[1:100], a$x[1:100], a$y[1:100], type = "h")
circos.link("a", 0, "b", 0, h = 0.3) #point to point
circos.link("c", c(-0.5, 0.5), "d", c(-0.5, 0.5), col = "red", border = NA, h = 0.2) #intreval to interval
circos.link("e", 0, "g", c(-1, 1), col = "green", border = "black", lwd = 2, lty = 2) #point to interval
circlize详述
circlize的绘图规则是初始化(initialize)-创建track-添加图形元素-创建track-添加图形元素-…-circos.clear。具体参数设置以及解释由于内容太多,有兴趣的可以自己参考文档。 我认为比较重要的是要理解track、sector。由于基本所有的图形元素我们都是添加在sector
里面,因此就需要指定track.index以及sector.index。接下来就用个例子来讲解一下如何操纵track、sector。
par(mar = c(1, 1, 1, 1))
factors <- letters[1:8]
circos.initialize(factors = factors, xlim = c(0, 1)) #初始化# 绘制三个track,并显示具体信息
for (i in 1:3) {
circos.trackPlotRegion(ylim = c(0, 1))}
circos.info(plot = TRUE)
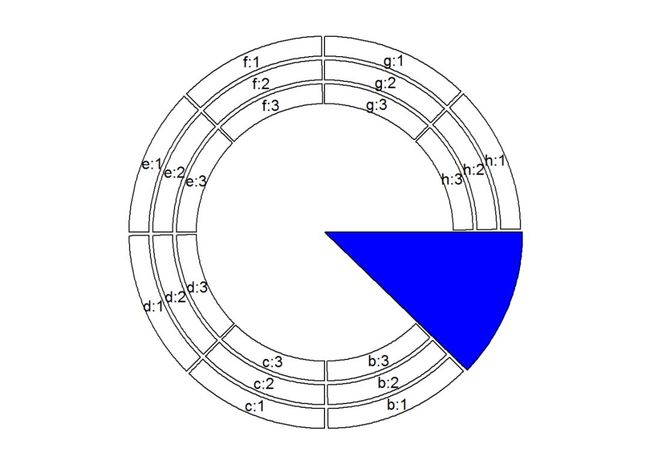
# 通过draw.sector()来高亮某一sector,比如a:
draw.sector(get.cell.meta.data("cell.start.degree", sector.index = "a"),
get.cell.meta.data("cell.end.degree", sector.index = "a"), rou1 = 1, col = "blue")
circos.clear()
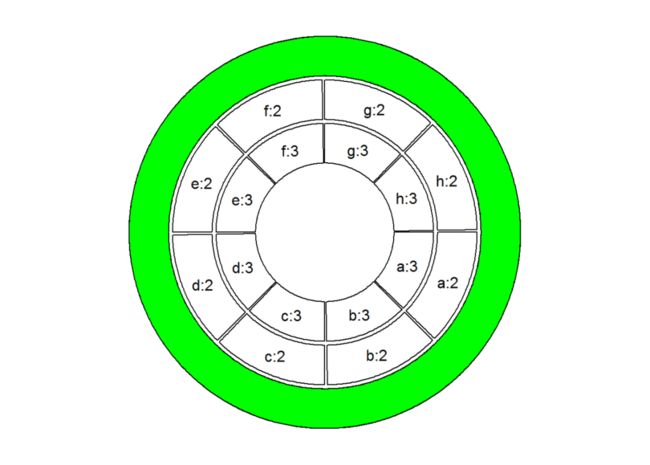
### 高亮某一track, 比如第一个track:
circos.initialize(factors = factors, xlim = c(0, 1))
for (i in 1:3) {
circos.trackPlotRegion(ylim = c(0, 1))}
circos.info(plot = TRUE)
draw.sector(0, 360, rou1 = get.cell.meta.data("cell.top.radius", track.index = 1),
rou2 = get.cell.meta.data("cell.bottom.radius", track.index = 1), col = "green")
circos.clear()
# 高亮某一track某一sector,比如地2、3track中的e、f(sector):
circos.initialize(factors = factors, xlim = c(0, 1))
for (i in 1:3) { circos.trackPlotRegion(ylim = c(0, 1))}
circos.info(plot = TRUE)
draw.sector(get.cell.meta.data("cell.start.degree", sector.index = "e"),
get.cell.meta.data("cell.end.degree", sector.index = "f"),
get.cell.meta.data("cell.top.radius", track.index = 2),
get.cell.meta.data("cell.bottom.radius", track.index = 3), col = "red")
circos.clear() #千万别忘了circos.clear,不然下次无法绘图。
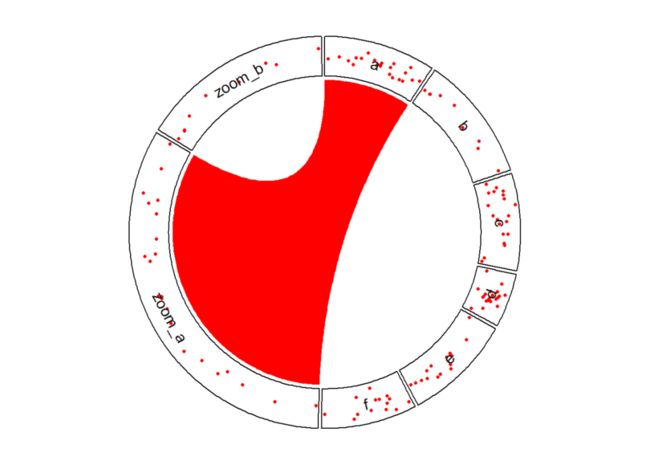
放大某一特定区域
df <- data.frame(factors = sample(letters[1:6], 100, replace = TRUE),
x = rnorm(100),
y = rnorm(100),
stringsAsFactors = FALSE)
# 放大a,b区域
zoom_df <- df %>% dplyr::filter(factors %in% c("a", "b"))
zoom_df$factors <- paste0("zoom_", zoom_df$factors)
df2 <- rbind(df, zoom_df)
xrange <- tapply(df2$x, df2$factors, function(x) max(x) - min(x))
normal_sector_index <- unique(df$factors)
zoomed_sector_index <- unique(zoom_df$factors)
sector.width <- c(xrange[normal_sector_index]/sum(xrange[normal_sector_index]),
xrange[zoomed_sector_index]/sum(xrange[zoomed_sector_index]))
# 绘图
par(mar = c(1, 1, 1, 1))
circos.par(start.degree = 90)
circos.initialize(df2$factors, x = df2$x, sector.width = sector.width)
circos.trackPlotRegion(df2$factors, x = df2$x, y = df2$y,
panel.fun = function(x, y) {
circos.points(x, y, col = "red", pch = 16, cex = 0.5)
xlim = get.cell.meta.data("xlim")
ylim = get.cell.meta.data("ylim")
sector.index = get.cell.meta.data("sector.index")
circos.text(mean(xlim), mean(ylim), sector.index, niceFacing = TRUE)})
# 添加links
circos.link("a", get.cell.meta.data("cell.xlim", sector.index = "a"), "zoom_a",
get.cell.meta.data("cell.xlim", sector.index = "zoom_a"), border = NA, col = "red")
circos.clear()
举个栗子
圈圈图+热图+进化树
set.seed(1234)
data <- matrix(rnorm(100 * 10), nrow = 10, ncol = 100)
col <- colorRamp2(c(-2, 0, 2), c("green", "black", "red"))
factors <- rep(letters[1:2], times = c(30, 70))
data_list <- list(a = data[, factors == "a"], b = data[, factors == "b"])
dend_list <- list(a = as.dendrogram(hclust(dist(t(data_list[["a"]])))),
b = as.dendrogram(hclust(dist(t(data_list[["b"]])))))
circos.par(cell.padding = c(0, 0, 0, 0), gap.degree = 5)
circos.initialize(factors = factors, xlim = cbind(c(0, 0), table(factors)))
circos.track(ylim = c(0, 10), bg.border = NA,
panel.fun = function(x, y) {
sector.index = get.cell.meta.data("sector.index")
d = data_list[[sector.index]]
dend = dend_list[[sector.index]]
d2 = d[, order.dendrogram(dend)]
col_data = col(d2)
nr = nrow(d2)
nc = ncol(d2)
for (i in 1:nr) {
circos.rect(1:nc - 1, rep(nr - i, nc), 1:nc, rep(nr - i + 1, nc),
border = col_data[i, ], col = col_data[i, ]) }})
max_height <- max(sapply(dend_list, function(x) attr(x, "height")))
circos.track(ylim = c(0, max_height),
bg.border = NA, track.height = 0.3,
panel.fun = function(x, y) {
sector.index = get.cell.meta.data("sector.index")
dend = dend_list[[sector.index]]
circos.dendrogram(dend, max_height = max_height)})
circos.clear()
多图排列
直接用layout设置
layout(matrix(1:9, 3, 3))
for (i in 1:9) {
factors = letters[1:8]
par(mar = c(0.5, 0.5, 0.5, 0.5))
circos.par(cell.padding = c(0, 0, 0, 0))
circos.initialize(factors = factors, xlim = c(0, 1))
circos.trackPlotRegion(ylim = c(0, 1), track.height = 0.05,
bg.col = rand_color(8), bg.border = NA)
# 绘制links
for (i in 1:20) {
se = sample(letters[1:8], 2)
circos.link(se[1], runif(2), se[2], runif(2),
col = rand_color(1, transparency = 0.4), border = NA)
}
circos.clear()
sessionInfo
理解circlize包的原理,绘制基因组数据的图形也是一样的。有时间下次介绍(主要是我自己还没看完,看不太懂)。
老规矩,给出sessionInfo。
sessionInfo()
## R version 3.4.0 (2017-04-21)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 8.1 x64 (build 9600)
## ## Matrix products: default##
## locale:
## [1] LC_COLLATE=Chinese (Simplified)_China.936
## [2] LC_CTYPE=Chinese (Simplified)_China.936
## [3] LC_MONETARY=Chinese (Simplified)_China.936
## [4] LC_NUMERIC=C
## [5] LC_TIME=Chinese (Simplified)_China.936
## ## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
## ## other attached packages:
## [1] circlize_0.4.0 BiocInstaller_1.26.0 forcats_0.2.0
## [4] stringr_1.2.0 dplyr_0.5.0 purrr_0.2.2.2
## [7] readr_1.1.1 tidyr_0.6.3 tibble_1.3.1
## [10] ggplot2_2.2.1 tidyverse_1.1.1.9000
#### loaded via a namespace (and not attached):
## [1] shape_1.4.2 clisymbols_1.2.0 reshape2_1.4.2
## [4] haven_1.0.0 lattice_0.20-35 colorspace_1.3-2
## [7] htmltools_0.3.6 yaml_2.1.14 rlang_0.1.1
## [10] foreign_0.8-68 DBI_0.6-1 modelr_0.1.0
## [13] readxl_1.0.0 plyr_1.8.4 munsell_0.4.3
## [16] gtable_0.2.0 cellranger_1.1.0 rvest_0.3.2
## [19] GlobalOptions_0.0.12 psych_1.7.5 evaluate_0.10
## [22] knitr_1.16 parallel_3.4.0 broom_0.4.2
## [25] Rcpp_0.12.11 scales_0.4.1 backports_1.1.0
## [28] formatR_1.5 jsonlite_1.4 boxes_0.0.0.9000
## [31] mnormt_1.5-5 hms_0.3 digest_0.6.12
## [34] stringi_1.1.5 grid_3.4.0 rprojroot_1.2
## [37] tools_3.4.0 magrittr_1.5 lazyeval_0.2.0
## [40] crayon_1.3.2.9000 xml2_1.1.1 lubridate_1.6.0
## [43] assertthat_0.2.0 rmarkdown_1.5 httr_1.2.1
## [46] rstudioapi_0.6 R6_2.2.1 nlme_3.1-131
## [49] compiler_3.4.0
猜你喜欢
10000+:肠道细菌 人体上的生命 宝宝与猫狗 梅毒狂想曲 提DNA发Nature 实验分析谁对结果影响大 Cell微生物专刊
系列教程:微生物组入门 Biostar 微生物组 宏基因组
专业技能:生信宝典 学术图表 高分文章 不可或缺的人
一文读懂:宏基因组 寄生虫益处 进化树
必备技能:提问 搜索 Endnote
文献阅读 热心肠 SemanticScholar Geenmedical
扩增子分析:图表解读 分析流程 统计绘图
16S功能预测 PICRUSt FAPROTAX Bugbase Tax4Fun
在线工具:16S预测培养基 生信绘图
科研经验:云笔记 云协作 公众号
编程模板 Shell R Perl
生物科普 生命大跃进 细胞暗战 人体奥秘
写在后面
为鼓励读者交流、快速解决科研困难,我们建立了“宏基因组”专业讨论群,目前己有国内外1700+ 一线科研人员加入。参与讨论,获得专业解答,欢迎分享此文至朋友圈,并扫码加主编好友带你入群,务必备注“姓名-单位-研究方向-职称/年级”。技术问题寻求帮助,首先阅读《如何优雅的提问》学习解决问题思路,仍末解决群内讨论,问题不私聊,帮助同行。
![]()
学习16S扩增子、宏基因组科研思路和分析实战,关注“宏基因组”
![]()
![]()