【实验小结】cs231n assignment1 knn 部分
1. 前言
这个是斯坦福 cs231n 课程的课程作业, 在做这个课程作业的过程中, 遇到了各种问题, 通过查阅资料加以解决, 加深了对课程内容的理解, 以及熟悉了相应的python 代码实现
工程地址: https://github.com/zhyh2010/cs231n/tree/master/assignment1
2. 具体实现部分
2.1 knn 调用程序
2.1.1 简单说明
- knn 算法原理非常简单, 我们之前也总结过一次: http://blog.csdn.net/zhyh1435589631/article/details/53875182
- 这个算法需要 对每个输入的测试数据计算他与所有的训练集数据之间的距离 (可以是 曼哈顿距离 L1, 欧式距离 L2), 然后挑选出其中距离最小的k个值作为 选民, 并根据他们的党派进行投票, 这是一种典型的少数服从多数的方法
2.1.2 knn 调用程序 代码分析
2.1.2.1 data_utils 载入数据集
- 这里选用的数据集是 cifar-10 数据集 http://www.cs.toronto.edu/~kriz/cifar.html
- 载入代码:
输出相应的训练集和测试集数据 Xtr, Ytr, Xte, Yte
def load_CIFAR_batch(filename):
""" load single batch of cifar """
with open(filename, 'rb') as f:
datadict = pickle.load(f)
X = datadict['data']
Y = datadict['labels']
X = X.reshape(10000, 3, 32, 32).transpose(0,2,3,1).astype("float")
Y = np.array(Y)
return X, Y
def load_CIFAR10(ROOT):
""" load all of cifar """
xs = []
ys = []
for b in range(1,6):
f = os.path.join(ROOT, 'data_batch_%d' % (b, ))
X, Y = load_CIFAR_batch(f)
xs.append(X)
ys.append(Y)
Xtr = np.concatenate(xs)
Ytr = np.concatenate(ys)
del X, Y
Xte, Yte = load_CIFAR_batch(os.path.join(ROOT, 'test_batch'))
return Xtr, Ytr, Xte, Yte2.1.2.2 载入数据集的调用
# Run some setup code for this notebook.
import random
import numpy as np
from cs231n.data_utils import load_CIFAR10
import matplotlib.pyplot as plt
# This is a bit of magic to make matplotlib figures appear inline in the notebook
# rather than in a new window.
%matplotlib inline
plt.rcParams['figure.figsize'] = (10.0, 8.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# Some more magic so that the notebook will reload external python modules;
# see http://stackoverflow.com/questions/1907993/autoreload-of-modules-in-ipython
%load_ext autoreload
%autoreload 2
# Load the raw CIFAR-10 data.
cifar10_dir = 'cs231n/datasets/cifar-10-batches-py'
X_train, y_train, X_test, y_test = load_CIFAR10(cifar10_dir)
# As a sanity check, we print out the size of the training and test data.
print 'Training data shape: ', X_train.shape
print 'Training labels shape: ', y_train.shape
print 'Test data shape: ', X_test.shape
print 'Test labels shape: ', y_test.shape显示结果:
Training data shape: (50000L, 32L, 32L, 3L)
Training labels shape: (50000L,)
Test data shape: (10000L, 32L, 32L, 3L)
Test labels shape: (10000L,)2.1.2.3 显示数据集的一部分信息
# Visualize some examples from the dataset.
# We show a few examples of training images from each class.
classes = ['plane', 'car', 'bird', 'cat', 'deer', 'dog', 'frog', 'horse', 'ship', 'truck']
num_classes = len(classes)
samples_per_class = 7
for y, cls in enumerate(classes):
idxs = np.flatnonzero(y_train == y)
idxs = np.random.choice(idxs, samples_per_class, replace=False)
for i, idx in enumerate(idxs):
plt_idx = i * num_classes + y + 1
plt.subplot(samples_per_class, num_classes, plt_idx)
plt.imshow(X_train[idx].astype('uint8'))
plt.axis('off')
if i == 0:
plt.title(cls)
plt.show()2.1.2.4 调整数据集大小
# Subsample the data for more efficient code execution in this exercise
num_training = 5000
mask = range(num_training)
X_train = X_train[mask]
y_train = y_train[mask]
num_test = 500
mask = range(num_test)
X_test = X_test[mask]
y_test = y_test[mask]
# Reshape the image data into rows
X_train = np.reshape(X_train, (X_train.shape[0], -1))
X_test = np.reshape(X_test, (X_test.shape[0], -1))
print X_train.shape, X_test.shape显示结果:
(5000L, 3072L) (500L, 3072L)2.1.2.5 使用KNN进行训练
这段 代码训练的时间特别长。。。。。
from cs231n.classifiers import KNearestNeighbor
# Create a kNN classifier instance.
# Remember that training a kNN classifier is a noop:
# the Classifier simply remembers the data and does no further processing
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
# Open cs231n/classifiers/k_nearest_neighbor.py and implement
# compute_distances_two_loops.
# Test your implementation:
dists = classifier.compute_distances_two_loops(X_test)
print dists.shape
# Now implement the function predict_labels and run the code below:
# We use k = 1 (which is Nearest Neighbor).
y_test_pred = classifier.predict_labels(dists, k=1)
# Compute and print the fraction of correctly predicted examples
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print 'Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy)显示结果:
Got 137 / 500 correct => accuracy: 0.2740002.1.2.6 修改 k 参数
y_test_pred = classifier.predict_labels(dists, k=5)
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print 'Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy)显示结果:
Got 145 / 500 correct => accuracy: 0.2900002.1.2.7 验证其他两种实现方式
结果是一致的
# Now lets speed up distance matrix computation by using partial vectorization
# with one loop. Implement the function compute_distances_one_loop and run the
# code below:
dists_one = classifier.compute_distances_one_loop(X_test)
# To ensure that our vectorized implementation is correct, we make sure that it
# agrees with the naive implementation. There are many ways to decide whether
# two matrices are similar; one of the simplest is the Frobenius norm. In case
# you haven't seen it before, the Frobenius norm of two matrices is the square
# root of the squared sum of differences of all elements; in other words, reshape
# the matrices into vectors and compute the Euclidean distance between them.
difference = np.linalg.norm(dists - dists_one, ord='fro')
print 'Difference was: %f' % (difference, )
if difference < 0.001:
print 'Good! The distance matrices are the same'
else:
print 'Uh-oh! The distance matrices are different'
# Now implement the fully vectorized version inside compute_distances_no_loops
# and run the code
dists_two = classifier.compute_distances_no_loops(X_test)
# check that the distance matrix agrees with the one we computed before:
difference = np.linalg.norm(dists - dists_two, ord='fro')
print 'Difference was: %f' % (difference, )
if difference < 0.001:
print 'Good! The distance matrices are the same'
else:
print 'Uh-oh! The distance matrices are different'2.1.2.8 查看三种实现方法的使用时间
# Let's compare how fast the implementations are
def time_function(f, *args):
"""
Call a function f with args and return the time (in seconds) that it took to execute.
"""
import time
tic = time.time()
f(*args)
toc = time.time()
return toc - tic
two_loop_time = time_function(classifier.compute_distances_two_loops, X_test)
print 'Two loop version took %f seconds' % two_loop_time
one_loop_time = time_function(classifier.compute_distances_one_loop, X_test)
print 'One loop version took %f seconds' % one_loop_time
no_loop_time = time_function(classifier.compute_distances_no_loops, X_test)
print 'No loop version took %f seconds' % no_loop_time
# you should see significantly faster performance with the fully vectorized implementation显示结果:
Two loop version took 46.657000 seconds
One loop version took 109.456000 seconds
No loop version took 1.205000 seconds可以发现这个效率差别真不是一心半点的, 尽量使用矩阵操作少用循环
2.2.3 knn 本质实现部分 代码分析
2.2.3.1 KNearestNeighbor 类整体分析
- 本质上, 这是一个类, 有多个成员函数构成, 用户调用的时候, 只需要调用
train和predict即可得到想要的预测数据 - 其中,
compute_distances_two_loops,compute_distances_one_loop,compute_distances_no_loops分别是用来实现需要预测的数据集X和 原始记录的训练集self.X_train之间的距离关系, 并通过predict_labels进行KNN预测
class KNearestNeighbor(object):
""" a kNN classifier with L2 distance """
def __init__(self):
pass
def train(self, X, y):
...
def predict(self, X, k=1, num_loops=0):
...
def compute_distances_two_loops(self, X):
...
def compute_distances_one_loop(self, X):
...
def compute_distances_no_loops(self, X):
...
def getNormMatrix(self, x, lines_num):
...
def predict_labels(self, dists, k=1):
...2.2.3.2 compute_distances_two_loops
这个函数主要通过两层 for 循环对计算测试集与训练集数据之间的欧式距离
d2(I1,I2)=∑p(Ip1−Ip2)2−−−−−−−−−−−√ d 2 ( I 1 , I 2 ) = ∑ p ( I 1 p − I 2 p ) 2
def compute_distances_two_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a nested loop over both the training data and the
test data.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data.
Returns:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
is the Euclidean distance between the ith test point and the jth training
point.
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
for j in xrange(num_train):
#####################################################################
# TODO: #
# Compute the l2 distance between the ith test point and the jth #
# training point, and store the result in dists[i, j]. You should #
# not use a loop over dimension. #
#####################################################################
dists[i, j] = np.sqrt(np.dot(X[i] - self.X_train[j], X[i] - self.X_train[j]))
#####################################################################
# END OF YOUR CODE #
#####################################################################
return dists2.2.3.3 compute_distances_one_loop
本质上这里填入的代码和 上一节中的是一致的, 只是多了一个 axis = 1 指定方向
def compute_distances_one_loop(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a single loop over the test data.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in xrange(num_test):
#######################################################################
# TODO: #
# Compute the l2 distance between the ith test point and all training #
# points, and store the result in dists[i, :]. #
#######################################################################
dists[i, :] = np.sqrt(np.sum(np.square(X[i] - self.X_train), axis = 1))
#######################################################################
# END OF YOUR CODE #
#######################################################################
return dists2.2.3.4 compute_distances_no_loops
- 这部分公式虽然短小, 但是需要一定的数学功底, 参考文章: http://blog.csdn.net/geekmanong/article/details/51524402
- 我们记测试集矩阵 为 P P 大小为 M×D M × D , 训练集矩阵 为 C C 大小为 N×D N × D
- 记 Pi P i 是 P P 的第 i i 行, 同理 Cj C j 是 C C 的 第 j j 行:
Pi=[Pi1Pi2⋯PiD]Cj=[Cj1Cj2⋯CjD] P i = [ P i 1 P i 2 ⋯ P i D ] C j = [ C j 1 C j 2 ⋯ C j D ] - 我们先来计算一下 Pi P i 和 Cj C j 之间的距离
d(Pi,Cj)=(Pi1−Pj1)2+(Pi2−Pj2)2+⋯+(PiD−PjD)2−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−√=(P2i1+P2i2+⋯+P2iD)+(C2j1+C2j2+⋯+C2jD)−2∗(Pi1Cj1+Pi2Cj2+⋯+PiDCjD)−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−√=||Pi||2+||Cj||2−2∗PiC′j−−−−−−−−−−−−−−−−−−−−√ d ( P i , C j ) = ( P i 1 − P j 1 ) 2 + ( P i 2 − P j 2 ) 2 + ⋯ + ( P i D − P j D ) 2 = ( P i 1 2 + P i 2 2 + ⋯ + P i D 2 ) + ( C j 1 2 + C j 2 2 + ⋯ + C j D 2 ) − 2 ∗ ( P i 1 C j 1 + P i 2 C j 2 + ⋯ + P i D C j D ) = | | P i | | 2 + | | C j | | 2 − 2 ∗ P i C j ′ - 我们可以推广得到,结果矩阵的每行元素为:
Res(i)=(||Pi||2||Pi||2⋯||Pi||2)+(||C1||2||C2||2⋯||CN||2)−2∗Pi(C′1C′2⋯C′N)−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−√=(||Pi||2||Pi||2⋯||Pi||2)+(||C1||2||C2||2⋯||CN||2)−2∗PiC′−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−√ R e s ( i ) = ( | | P i | | 2 | | P i | | 2 ⋯ | | P i | | 2 ) + ( | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 ) − 2 ∗ P i ( C 1 ′ C 2 ′ ⋯ C N ′ ) = ( | | P i | | 2 | | P i | | 2 ⋯ | | P i | | 2 ) + ( | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 ) − 2 ∗ P i C ′ - 继而, 结果矩阵为:
Res=⎛⎝⎜⎜⎜⎜⎜⎜||P1||2||P2||2⋮||PM||2||P1||2||P2||2⋮||PM||2⋯⋯⋱⋯||P1||2||P2||2⋮||PM||2⎞⎠⎟⎟⎟⎟⎟⎟+⎛⎝⎜⎜⎜⎜⎜⎜||C1||2||C1||2⋮||C1||2||C2||2||C2||2⋮||C2||2⋯⋯⋱⋯||CN||2||CN||2⋮||CN||2⎞⎠⎟⎟⎟⎟⎟⎟−2PC′−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−⎷=⎛⎝⎜⎜⎜⎜⎜||P1||2||P2||2⋮||PM||2⎞⎠⎟⎟⎟⎟⎟M×1∗(11⋯1)1×N+⎛⎝⎜⎜⎜⎜11⋮1⎞⎠⎟⎟⎟⎟M×1∗(||C1||2||C2||2⋯||CN||2)1×N−2PM×DC′N×D−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−−⎷ R e s = ( | | P 1 | | 2 | | P 1 | | 2 ⋯ | | P 1 | | 2 | | P 2 | | 2 | | P 2 | | 2 ⋯ | | P 2 | | 2 ⋮ ⋮ ⋱ ⋮ | | P M | | 2 | | P M | | 2 ⋯ | | P M | | 2 ) + ( | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 ⋮ ⋮ ⋱ ⋮ | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 ) − 2 P C ′ = ( | | P 1 | | 2 | | P 2 | | 2 ⋮ | | P M | | 2 ) M × 1 ∗ ( 1 1 ⋯ 1 ) 1 × N + ( 1 1 ⋮ 1 ) M × 1 ∗ ( | | C 1 | | 2 | | C 2 | | 2 ⋯ | | C N | | 2 ) 1 × N − 2 P M × D C N × D ′ - 转换为python 代码如下:
def compute_distances_no_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using no explicit loops.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
#########################################################################
# TODO: #
# Compute the l2 distance between all test points and all training #
# points without using any explicit loops, and store the result in #
# dists. #
# #
# You should implement this function using only basic array operations; #
# in particular you should not use functions from scipy. #
# #
# HINT: Try to formulate the l2 distance using matrix multiplication #
# and two broadcast sums. #
#########################################################################
dists = np.sqrt(self.getNormMatrix(X, num_train).T + self.getNormMatrix(self.X_train, num_test) - 2 * np.dot(X, self.X_train.T))
#########################################################################
# END OF YOUR CODE #
#########################################################################
return dists
def getNormMatrix(self, x, lines_num):
"""
Get a lines_num x size(x, 1) matrix
"""
return np.ones((lines_num, 1)) * np.sum(np.square(x), axis = 1) 从最终得到的结果看, 这个推导的结果运行速度是最快的
2.2.3.5 predict_labels
根据计算得到的距离关系, 挑选 K 个数据组成选民, 进行党派选举
def predict_labels(self, dists, k=1):
"""
Given a matrix of distances between test points and training points,
predict a label for each test point.
Inputs:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
gives the distance betwen the ith test point and the jth training point.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
num_test = dists.shape[0]
y_pred = np.zeros(num_test)
for i in xrange(num_test):
# A list of length k storing the labels of the k nearest neighbors to
# the ith test point.
closest_y = []
#########################################################################
# TODO: #
# Use the distance matrix to find the k nearest neighbors of the ith #
# testing point, and use self.y_train to find the labels of these #
# neighbors. Store these labels in closest_y. #
# Hint: Look up the function numpy.argsort. #
#########################################################################
kids = np.argsort(dists[i])
closest_y = self.y_train[kids[:k]]
#########################################################################
# TODO: #
# Now that you have found the labels of the k nearest neighbors, you #
# need to find the most common label in the list closest_y of labels. #
# Store this label in y_pred[i]. Break ties by choosing the smaller #
# label. #
#########################################################################
count = 0
label = 0
for j in closest_y:
tmp = 0
for kk in closest_y:
tmp += (kk == j)
if tmp > count:
count = tmp
label = j
y_pred[i] = label
#y_pred[i] = np.argmax(np.bincount(closest_y))
#########################################################################
# END OF YOUR CODE #
#########################################################################
return y_pred2.2.3.6 predict
- 这里主要做了两个步骤:
- 计算欧式距离
- KNN 统计预测信息
def predict(self, X, k=1, num_loops=0):
"""
Predict labels for test data using this classifier.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data consisting
of num_test samples each of dimension D.
- k: The number of nearest neighbors that vote for the predicted labels.
- num_loops: Determines which implementation to use to compute distances
between training points and testing points.
Returns:
- y: A numpy array of shape (num_test,) containing predicted labels for the
test data, where y[i] is the predicted label for the test point X[i].
"""
if num_loops == 0:
dists = self.compute_distances_no_loops(X)
elif num_loops == 1:
dists = self.compute_distances_one_loop(X)
elif num_loops == 2:
dists = self.compute_distances_two_loops(X)
else:
raise ValueError('Invalid value %d for num_loops' % num_loops)
return self.predict_labels(dists, k=k)2.2.4 cross-validation 代码分析
- 交叉验证实际上是将数据的训练集进行拆分, 分成多个组, 构成多个训练和测试集, 来筛选较好的超参数
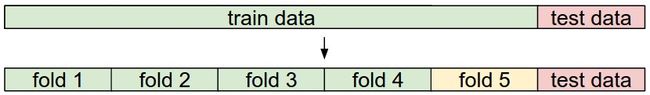
- 如图所示, 可以分为 5组数据, (分别将 fold 1, 2 .. 5 作为验证集, 将剩余的数据作为训练集, 训练得到超参数)
2.2.4.1 筛选不同的k
num_folds = 5
k_choices = [1, 3, 5, 8, 10, 12, 15, 20, 50, 100]
X_train_folds = []
y_train_folds = []
################################################################################
# TODO: #
# Split up the training data into folds. After splitting, X_train_folds and #
# y_train_folds should each be lists of length num_folds, where #
# y_train_folds[i] is the label vector for the points in X_train_folds[i]. #
# Hint: Look up the numpy array_split function. #
################################################################################
X_train_folds = np.array_split(X_train, num_folds)
y_train_folds = np.array_split(y_train, num_folds)
################################################################################
# END OF YOUR CODE #
################################################################################
# A dictionary holding the accuracies for different values of k that we find
# when running cross-validation. After running cross-validation,
# k_to_accuracies[k] should be a list of length num_folds giving the different
# accuracy values that we found when using that value of k.
k_to_accuracies = {}
################################################################################
# TODO: #
# Perform k-fold cross validation to find the best value of k. For each #
# possible value of k, run the k-nearest-neighbor algorithm num_folds times, #
# where in each case you use all but one of the folds as training data and the #
# last fold as a validation set. Store the accuracies for all fold and all #
# values of k in the k_to_accuracies dictionary. #
################################################################################
for k in k_choices:
k_to_accuracies[k] = np.zeros(num_folds)
for i in range(num_folds):
Xtr = np.array(X_train_folds[:i] + X_train_folds[i+1:])
ytr = np.array(y_train_folds[:i] + y_train_folds[i+1:])
Xte = np.array(X_train_folds[i])
yte = np.array(y_train_folds[i])
Xtr = np.reshape(Xtr, (X_train.shape[0] * 4 / 5, -1))
ytr = np.reshape(ytr, (y_train.shape[0] * 4 / 5, -1))
Xte = np.reshape(Xte, (X_train.shape[0] / 5, -1))
yte = np.reshape(yte, (y_train.shape[0] / 5, -1))
classifier.train(Xtr, ytr)
yte_pred = classifier.predict(Xte, k)
yte_pred = np.reshape(yte_pred, (yte_pred.shape[0], -1))
num_correct = np.sum(yte_pred == yte)
accuracy = float(num_correct) / len(yte)
k_to_accuracies[k][i] = accuracy
################################################################################
# END OF YOUR CODE #
################################################################################
# Print out the computed accuracies
for k in sorted(k_to_accuracies):
for accuracy in k_to_accuracies[k]:
print 'k = %d, accuracy = %f' % (k, accuracy)显示结果:
k = 1, accuracy = 0.263000
k = 1, accuracy = 0.257000
k = 1, accuracy = 0.264000
k = 1, accuracy = 0.278000
k = 1, accuracy = 0.266000
k = 3, accuracy = 0.257000
k = 3, accuracy = 0.263000
k = 3, accuracy = 0.273000
k = 3, accuracy = 0.282000
k = 3, accuracy = 0.270000
k = 5, accuracy = 0.265000
k = 5, accuracy = 0.275000
k = 5, accuracy = 0.295000
k = 5, accuracy = 0.298000
k = 5, accuracy = 0.284000
k = 8, accuracy = 0.272000
k = 8, accuracy = 0.295000
k = 8, accuracy = 0.284000
k = 8, accuracy = 0.298000
k = 8, accuracy = 0.290000
k = 10, accuracy = 0.272000
k = 10, accuracy = 0.303000
k = 10, accuracy = 0.289000
k = 10, accuracy = 0.292000
k = 10, accuracy = 0.285000
k = 12, accuracy = 0.271000
k = 12, accuracy = 0.305000
k = 12, accuracy = 0.285000
k = 12, accuracy = 0.289000
k = 12, accuracy = 0.281000
k = 15, accuracy = 0.260000
k = 15, accuracy = 0.302000
k = 15, accuracy = 0.292000
k = 15, accuracy = 0.292000
k = 15, accuracy = 0.285000
k = 20, accuracy = 0.268000
k = 20, accuracy = 0.293000
k = 20, accuracy = 0.291000
k = 20, accuracy = 0.287000
k = 20, accuracy = 0.286000
k = 50, accuracy = 0.273000
k = 50, accuracy = 0.291000
k = 50, accuracy = 0.274000
k = 50, accuracy = 0.267000
k = 50, accuracy = 0.273000
k = 100, accuracy = 0.261000
k = 100, accuracy = 0.272000
k = 100, accuracy = 0.267000
k = 100, accuracy = 0.260000
k = 100, accuracy = 0.2670002.2.4.2 图形化显示
# plot the raw observations
for k in k_choices:
accuracies = k_to_accuracies[k]
plt.scatter([k] * len(accuracies), accuracies)
# plot the trend line with error bars that correspond to standard deviation
accuracies_mean = np.array([np.mean(v) for k,v in sorted(k_to_accuracies.items())])
accuracies_std = np.array([np.std(v) for k,v in sorted(k_to_accuracies.items())])
plt.errorbar(k_choices, accuracies_mean, yerr=accuracies_std)
plt.title('Cross-validation on k')
plt.xlabel('k')
plt.ylabel('Cross-validation accuracy')
plt.show()2.2.4.3 选取最好的k 进行训练
# Based on the cross-validation results above, choose the best value for k,
# retrain the classifier using all the training data, and test it on the test
# data. You should be able to get above 28% accuracy on the test data.
best_k = 8
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
y_test_pred = classifier.predict(X_test, k=best_k)
# Compute and display the accuracy
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print 'Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy)显示结果:
Got 147 / 500 correct => accuracy: 0.294000可以发现, 即使是最好情况下, KNN算法的识别准确率也只有30%, 因而, 一般不用来做图像分类
3. 参考资料
- 寒老师博客 http://blog.csdn.net/han_xiaoyang/article/details/49949535
- cs231n 课程主页 http://vision.stanford.edu/teaching/cs231n/syllabus.html
- github 主页 http://cs231n.github.io/
- http://www.cnblogs.com/daihengchen/p/5754383.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.sqrt.html
- https://docs.scipy.org/doc/numpy-1.10.1/reference/generated/numpy.sum.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.ndarray.shape.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.reshape.html
- enumerate : http://blog.chinaunix.net/uid-27040911-id-3429751.html
- https://docs.scipy.org/doc/numpy-dev/reference/generated/numpy.random.choice.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.flatnonzero.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.sum.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.concatenate.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.ones.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.count_nonzero.html#numpy.count_nonzero
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.argsort.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.bincount.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.multiply.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.dot.html
- https://docs.scipy.org/doc/numpy/reference/generated/numpy.array_split.html

